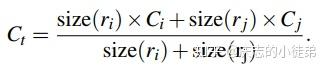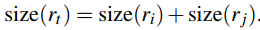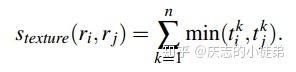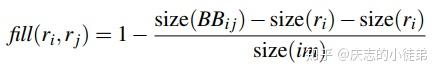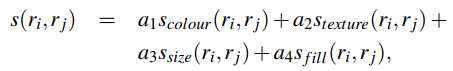深度学习基础
https://gombru.github.io/2018/05/23/cross_entropy_loss/
https://www.zhihu.com/question/382802283/answer/1114719159
https://zhuanlan.zhihu.com/p/93658728
Droupout
https://zhuanlan.zhihu.com/p/146747803
一、评估指标
代价函数:f(\theta,y),又称Cost function,loss function objective function。一般用在训练过程中,用来定义预测值和真实值之间的距离(也就是衡量模型在训练集上的性能),作为模型调整参数的反馈。代价函数越小,模型性能越好。
评判指标:f(\hat y,y),一般用于训练和测试过程中,用于评估模型好坏。评判指标越大(或越小),模型越好。
本质上代价函数和评判指标都是一家人,只他们的应用场景不同,分工不同。代价函数是用来优化模型参数的,评价指标是用来评判模型好坏的。
作为代价函数所具备的条件:
函数光滑且可导:可用梯度下降求解极值
函数为凸函数:可用梯度下降求解最优解
......
例如我们经常使用的分类器评判指标AUC就不能直接被优化,因此我们常采用交叉熵来代替AUC进行优化。一般情况下,交叉熵越小,AUC就会越大。
1.1 回归(Regression)算法指标
- 平均绝对误差(Mean Absolute Error)
平均绝对误差MAE(Mean Absolute Error)又被称为l1范数损失(l1-norm loss):
MAE不足:MAE虽能较好衡量回归模型的好坏,但是绝对值的存在导致函数不光滑,在某些点上不能求导,可以考虑将绝对值改为残差的平方,这就是均方误差。
def mean_absolute_error(y_true, y_pred, *, sample_weight=None, multioutput='uniform_average'):
"""Mean absolute error regression loss
Parameters
y_true : array-like of shape (n_samples,) or (n_samples, n_outputs)
Ground truth (correct) target values.
y_pred : array-like of shape (n_samples,) or (n_samples, n_outputs)
Estimated target values.
sample_weight : array-like of shape (n_samples,), optional
Sample weights.
multioutput : string in ['raw_values', 'uniform_average'] or array-like of shape (n_outputs)
Defines aggregating of multiple output values.
'raw_values' : Returns a full set of errors in case of multioutput input.
'uniform_average' : Errors of all outputs are averaged with uniform weight.
Returns
loss : float or ndarray of floats
If multioutput is 'raw_values', then mean absolute error is returned for each output separately.
If multioutput is 'uniform_average' or an ndarray of weights, then the weighted average of all output errors is returned.
MAE output is non-negative floating point. The best value is 0.0.
Examples
>>> from sklearn.metrics import mean_absolute_error
>>> y_true = [3, -0.5, 2, 7]
>>> y_pred = [2.5, 0.0, 2, 8]
>>> mean_absolute_error(y_true, y_pred)
0.5
>>> y_true = [[0.5, 1], [-1, 1], [7, -6]]
>>> y_pred = [[0, 2], [-1, 2], [8, -5]]
>>> mean_absolute_error(y_true, y_pred)
0.75
>>> mean_absolute_error(y_true, y_pred, multioutput='raw_values')
array([0.5, 1. ])
>>> mean_absolute_error(y_true, y_pred, multioutput=[0.3, 0.7])
0.85...
"""
y_type, y_true, y_pred, multioutput = _check_reg_targets(y_true, y_pred, multioutput)
check_consistent_length(y_true, y_pred, sample_weight)
output_errors = np.average(np.abs(y_pred - y_true), weights=sample_weight, axis=0)
if isinstance(multioutput, str):
if multioutput == 'raw_values':
return output_errors
elif multioutput == 'uniform_average':
# pass None as weights to np.average: uniform mean
multioutput = None
return np.average(output_errors, weights=multioutput)
- 均方误差(Mean Squared Error)
均方误差MSE(Mean Squared Error)又被称为l2范数损失(l2-norm loss):
MSE和方差的性质比较类似,与我们的目标变量的量纲不一致,为了保证量纲一致性,我们需要对MSE进行开方得到RMSE。
- 均方根误差(Root Mean Squared Error)
开方之后的MSE称为RMSE,是标准差的表兄弟,如下式所示:
上面的几种衡量标准的取值大小与具体的应用场景有关系,很难定义统一的规则来衡量模型的好坏。比如说利用机器学习算法预测上海的房价RMSE在2000元,我们是可以接受的,但是当四五线城市的房价RMSE为2000元,我们还可以接受吗?
def mean_squared_error(y_true, y_pred, *, sample_weight=None,multioutput='uniform_average', squared=True):
"""Mean squared error regression loss
Parameters
y_true : array-like of shape (n_samples,) or (n_samples, n_outputs)
Ground truth (correct) target values.
y_pred : array-like of shape (n_samples,) or (n_samples, n_outputs)
Estimated target values.
sample_weight : array-like of shape (n_samples,), optional
Sample weights.
multioutput : string in ['raw_values', 'uniform_average'] or array-like of shape (n_outputs)
Defines aggregating of multiple output values.
Array-like value defines weights used to average errors.
'raw_values' : Returns a full set of errors in case of multioutput input.
'uniform_average' : Errors of all outputs are averaged with uniform weight.
squared : boolean value, optional (default = True)
If True returns MSE value, if False returns RMSE value.
Returns
loss : float or ndarray of floats
A non-negative floating point value (the best value is 0.0), or an array of floating point values, one for each individual target.
Examples
>>> from sklearn.metrics import mean_squared_error
>>> y_true = [3, -0.5, 2, 7]
>>> y_pred = [2.5, 0.0, 2, 8]
>>> mean_squared_error(y_true, y_pred)
0.375
>>> y_true = [3, -0.5, 2, 7]
>>> y_pred = [2.5, 0.0, 2, 8]
>>> mean_squared_error(y_true, y_pred, squared=False)
0.612...
>>> y_true = [[0.5, 1],[-1, 1],[7, -6]]
>>> y_pred = [[0, 2],[-1, 2],[8, -5]]
>>> mean_squared_error(y_true, y_pred)
0.708...
>>> mean_squared_error(y_true, y_pred, multioutput='raw_values')
array([0.41666667, 1. ])
>>> mean_squared_error(y_true, y_pred, multioutput=[0.3, 0.7])
0.825...
"""
y_type, y_true, y_pred, multioutput = _check_reg_targets(y_true, y_pred, multioutput)
check_consistent_length(y_true, y_pred, sample_weight)
output_errors = np.average((y_true - y_pred) ** 2, axis=0, weights=sample_weight)
if isinstance(multioutput, str):
if multioutput == 'raw_values':
return output_errors if squared else np.sqrt(output_errors)
elif multioutput == 'uniform_average':
# pass None as weights to np.average: uniform mean
multioutput = None
mse = np.average(output_errors, weights=multioutput)
return mse if squared else np.sqrt(mse)
- 决定系数(Coefficient of determination)
变量之所以有价值,就是因为变量是变化的。什么意思呢?比如说一组因变量为[0,0,0,0,0],显然该因变量的结果是一个常数0,我们也没有必要建模对该因变量进行预测。假如一组的因变量为[1,3,7,10,12],该因变量是变化的,也就是有变异,因此需要通过建立回归模型进行预测。这里的变异可以理解为一组数据的方差不为0。
决定系数又称为R^2 score,反应因变量的全部变异能通过回归关系被自变量解释的比例。
如果结果是0,就说明模型预测不能预测因变量。如果结果是1。就说明是函数关系。如果结果是0~1之间的数,就是我们模型的好坏程度。化简上面的公式,分子就变成了我们的均方误差MSE,下面分母就变成了方差:
以上的评估指标是基于误差的均值对进行评估的,均值对异常点(outliers)较敏感,如果样本中有一些异常值出现,会对以上指标的值有较大影响,即均值是非鲁棒的。
- 解决评估指标鲁棒性问题
我们通常用一下两种方法解决评估指标的鲁棒性问题:
剔除异常值:设定一个相对误差\frac{|y_i-\hat{y_i}|}{y_i},当该值超过一定的阈值时,则认为其是一个异常点,剔除这个异常点,将异常点剔除之后。再计算平均误差来对模型进行评价。
使用误差的分位数来代替:如利用中位数来代替平均数。例如MAPE:MAPE=median(|y_i-\hat{y_i}|/y_i),MAPE是一个相对误差的中位数,当然也可以使用别的分位数。
https://zhuanlan.zhihu.com/p/38529433
1.2 常见的距离
在机器学习里,我们的运算一般都是基于向量的,一条用户具有100个特征,那么他对应的就是一个100维的向量,通过计算两个用户对应向量之间的距离值大小,有时候能反映出这两个用户的相似程度。这在后面的KNN算法和K-means算法中很明显。
一般而言,定义一个距离函数d(x,y),需要满足下面几个准则:
d(x,x)=0,到自己的距离为0
d(x,y)>= 0,距离非负
d(x,y)=d(y,x),对称性,如果A到B距离是a,那么B到A的距离也应该是a
d(x,k)+d(k,y)>= d(x,y),三角形法则:(两边之和大于第三边)
设有两个n维变量A=\left[ x_{11}, x_{12},...,x_{1n} \right]和B=\left[ x_{21} ,x_{22} ,...,x_{2n} \right],则一些常用的距离公式定义如下:
- 曼哈顿距离
曼哈顿距离也称为城市街区距离,数学定义如下:
曼哈顿距离的python实现:
from numpy import *
vector1 = mat([1,2,3])
vector2 = mat([4,5,6])
print(sum(abs(vector1-vector2)))
- 欧氏距离
欧氏距离其实就是L2范数,数学定义如下:
欧氏距离的Python实现:
from numpy import *
vector1 = mat([1,2,3])
vector2 = mat([4,5,6])
print(sqrt((vector1-vector2)*(vector1-vector2).T))
- 切比雪夫距离
切比雪夫距离就是L_{\infty},即无穷范数,数学表达式如下:
切比雪夫距离额Python实现如下:
from numpy import *
vector1 = mat([1,2,3])
vector2 = mat([4,5,6])
print(sqrt(abs(vector1-vector2).max))
- 闵可夫斯基距离
从严格意义上讲,闵可夫斯基距离不是一种距离,而是一组距离的定义:
该距离最常用的p是2和1,前者是欧几里得距离(Euclidean distance),后者是曼哈顿距离(Manhattan distance)。假设在曼哈顿街区乘坐出租车从P点到Q点,白色表示高楼大厦,灰色表示街道:

绿色的斜线表示欧几里得距离,在现实中是不可能的。其他三条折线表示了曼哈顿距离,这三条折线的长度是相等的。当p趋近于无穷大时,闵可夫斯基距离转化成切比雪夫距离(Chebyshev distance)。
我们知道平面上到原点欧几里得距离(p=2)为1的点所组成的形状是一个圆,当p取其他数值的时候呢?

注意,当p<1时,闵可夫斯基距离不再符合三角形法则,举个例子:当p<1,(0,0)到(1,1)的距离等于(1+1)^{1/p}>2,而(0,1)到这两个点的距离都是1。
闵可夫斯基距离比较直观,但是它与数据的分布有关,具有一定的局限性,如果x方向的幅值远远大于y方向的值,这个距离公式就会过度放大x维度的作用。所以,在计算距离之前,我们可能还需要对数据进行z-transform 处理,即减去均值,除以标准差:
其中\mu为该维度上的均值,\sigma为该维度上的标准差。
可以看到,上述处理开始体现数据的统计特性了。这种方法在假设数据各个维度不相关的情况下利用数据分布的特性计算出不同的距离。如果维度相互之间数据相关(例如:身高较高的信息很有可能会带来体重较重的信息,因为两者是有关联的),这时候就要用到马氏距离(Mahalanobis distance)了。
可以看到,上述处理开始体现数据的统计特性了。这种方法在假设数据各个维度不相关的情况下利用数据分布的特性计算出不同的距离。如果维度相互之间数据相关(例如:身高较高的信息很有可能会带来体重较重的信息,因为两者是有关联的),这时候就要用到马氏距离(Mahalanobis distance)了。
- 马氏距离
马氏距离实际上是利用Cholesky transformation来消除不同维度之间的相关性和尺度不同的性质。假设样本点(列向量)之间的协方差对称矩阵是\Sigma, 通过Cholesky Decomposition(实际上是对称矩阵LU分解的一种特殊形式)可以转化为下三角矩阵和上三角矩阵的乘积:\Sigma=LL^T。消除不同维度之间的相关性和尺度不同,只需要对样本点x做如下处理:z=L^{-1}(x-\mu)。处理之后的欧几里得距离就是原样本的马氏距离):
马氏距离的问题:
协方差矩阵必须满秩:里面有求逆矩阵的过程,不满秩不行,要求数据要有原维度个特征值,如果没有可以考虑先进行PCA,这种情况下PCA不会损失信息
不能处理非线性流形(manifold)上的问题:只对线性空间有效,如果要处理流形,只能在局部定义,可以用来建立KNN图
python代码:
import numpy as np
import pylab as pl
import scipy.spatial.distance as dist
def plotSamples(x, y, z=None):
stars = np.matrix([[3., -2., 0.], [3., 2., 0.]])
if z is not None:
x, y = z * np.matrix([x, y])
stars = z * stars
pl.scatter(x, y, s=10) # 画 gaussian 随机点
pl.scatter(np.array(stars[0]), np.array(stars[1]), s=200, marker='*', color='r') # 画三个指定点
pl.axhline(linewidth=2, color='g') # 画 x 轴
pl.axvline(linewidth=2, color='g') # 画 y 轴
pl.axis('equal')
pl.axis([-5, 5, -5, 5])
pl.show()
# 产生高斯分布的随机点
mean = [0, 0] # 平均值
cov = [[2, 1], [1, 2]] # 协方差
x, y = np.random.multivariate_normal(mean, cov, 1000).T
plotSamples(x, y)
covMat = np.matrix(np.cov(x, y)) # 求 x 与 y 的协方差矩阵
Z = np.linalg.cholesky(covMat).I # 仿射矩阵
plotSamples(x, y, Z)
# 求马氏距离
print('\n到原点的马氏距离分别是:')
print(dist.mahalanobis([0,0], [3,3], covMat.I), dist.mahalanobis([0,0], [-2,2], covMat.I))
# 求变换后的欧几里得距离
dots = (Z * np.matrix([[3, -2, 0], [3, 2, 0]])).T
print('\n变换后到原点的欧几里得距离分别是:')
print(dist.minkowski([0, 0], np.array(dots[0]), 2), dist.minkowski([0, 0], np.array(dots[1]), 2))
- 夹角余弦
夹角余弦的取值范围为[-1,1],可以用来衡量两个向量方向的差异;夹角余弦越大,表示两个向量的夹角越小;当两个向量的方向重合时,夹角余弦取最大值1;当两个向量的方向完全相反时,夹角余弦取最小值-1。
机器学习中用这一概念来衡量样本向量之间的差异,其数学表达式如下:
夹角余弦的python实现:
from numpy import *
vector1 = mat([1,2,3])
vector2 = mat([4,5,6])
print(dot(vector1,vector2)/(linalg.norm(vector1)*linalg.norm(vector2)))
- 汉明距离
汉明距离定义的是两个字符串中不相同位数的数目。例如:字符串‘1111’与‘1001’之间的汉明距离为2。信息编码中一般应使得编码间的汉明距离尽可能的小。
汉明距离的python实现:
from numpy import *
matV = mat([1,1,1,1],[1,0,0,1])
smstr = nonzero(matV[0]-matV[1])
print(smstr)
- 杰卡德相似系数
两个集合A和B的交集元素在A和B的并集中所占的比例称为两个集合的杰卡德相似系数,用符号J(A,B)表示,数学表达式为:
杰卡德相似系数是衡量两个集合的相似度的一种指标。一般可以将其用在衡量样本的相似度上。
- 杰卡德距离
与杰卡德相似系数相反的概念是杰卡德距离,其定义式为:
杰卡德距离的python实现:
from numpy import *
import scipy.spatial.distance as dist
matV = mat([1,1,1,1],[1,0,0,1])
print(dist.pdist(matV,'jaccard'))
- KL散度
KL散度不对称。如果分布P和Q,KL(P||Q)很大而KL(Q||P)很小
1.3 分类(Classification)指标
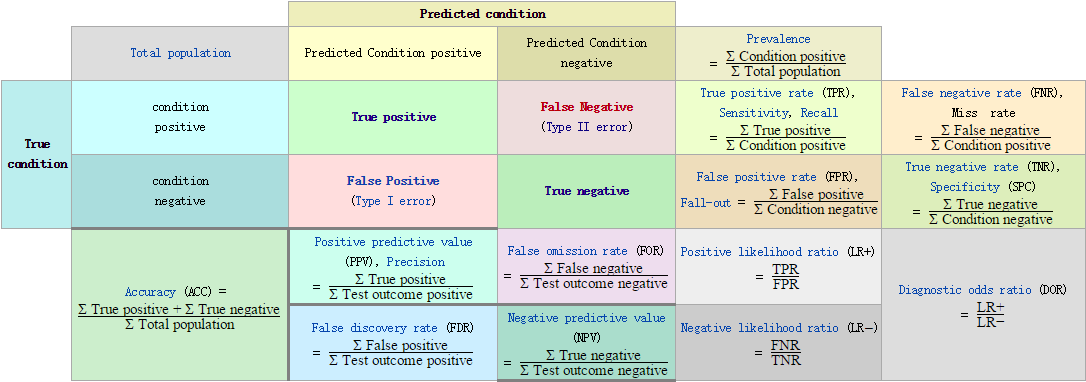
- 混淆矩阵
在预测系统中,牵扯到预测值,真实值,以及真实值和预测值之间的关系,进而产生了混淆矩阵:
 |
 |
| 多分类下的混淆矩阵 | 二分类下的混淆矩阵 |
混淆矩阵又被称为错误矩阵,在每个类别下,模型预测错误的结果数量,以及错误预测的类别和正确预测的数量都在一个矩阵下面显示出来,方便直观的评估模型分类的结果。
通常取预测值和真实值之间的关系、预测值对矩阵进行划分:
True positive(TP):真实值为Positive,预测正确(预测值为Positive)
True negative(TN):真实值为Negative,预测正确(预测值为Negative)
False positive(FP):真实值为Negative,预测错误(预测值为Positive),第一类错误,Type I error。
False negative(FN): 真实值为Positive,预测错误(预测值为 Negative),第二类错误,Type II error。
- 精确率(Precision)
精确率是针对我们预测结果而言的,它表示的是预测为正的样本中有多少是真正的正样本。那么预测为正就有两种可能了,一种就是把正类预测为正类(TP),另一种就是把负类预测为正类(FP),也就是
精确率取值范围为[0,1],取值越大,模型预测能力越好。
- 召回率(Recall)
针对我们原来的样本而言的,它表示的是样本中的正例有多少被预测正确了。那也有两种可能,一种是把原来的正类预测成正类(TP),另一种就是把原来的正类预测为负类(FN)。其实就是分母不同,一个分母是预测为正的样本数,另一个是原来样本中所有的正样本数。
在信息检索领域,精确率和召回率又被称为查准率和查全率:
查准率=检索出的相关信息量 / 检索出的信息总量
查全率=检索出的相关信息量 / 系统中的相关信息总量
- 准确率(Accuracy)
针对所有的样本,样本预测正确的数量占总数据:
- 引申指标
用样本中的正类和负类进行计算的定义
| 缩写 | 全称 | 等价称呼 | 计算公式 |
|---|---|---|---|
| TPR | True Positive Rate | 真正类率,Recall Sensitivity | \frac{TP}{TP+FN} |
| FNR | False Negative Rate | 假负类率,Miss rate Type rs error | \frac{TN}{TP+FN} |
| FPR | False Positive Rate | 假正类率,fall-out Type 1 error | \frac{FP}{FP+FN}=1-TNR |
| TNR | True Negative Rate | 真负类率,Specificity | \frac{TN}{TN+FP} |
用预测结果的正类和负类进行计算的定义
| 缩写 | 全称 | 等价称呼 | 计算公式 |
|---|---|---|---|
| PPV | Positive Predictive Value | 正类预测率,Precision | \frac{TP}{TP+FP} |
| FOR | False Omission Rata | 假错误率 | \frac{FN}{TN+FN}=1-NPV |
| FDR | False Discovery Rate | 假发现率 | \frac{FP}{TP+FP} |
| NPV | Negative Predictive Value | 负类预测率 | \frac{TN}{TN+FN} |
其他定义概念
| 缩写 | 全称 | 等价称呼 | 计算公式 |
|---|---|---|---|
| ACC | Accuracy | 准确率 | \frac{TP+TN}{TP+FN+FP+TN} |
| LR+ | Positive Likelihood Ratio | 正类似然比 | \frac{TPR}{FPR} |
| LR- | Negative likelihood ratio | 负类似然比 | \frac{FNR}{TNR} |
| DOR | Diagnostic odds ratio | 诊断胜算比 | \frac{LR+}{LR-} |
| F1 score | F1 test measure | F1值 | \frac{2*Recall*Precision}{Recall+Precision} |
| MCC | Matthews Correlation coefficient | 马修斯相关性系数 | \frac{TP*TN-FP*FN}{\sqrt{(TP+FP)(TP+FN)(TN+FP)(TN+FN)}} |
| BM | Bookmaker Informedness | Informedness | TPR+TNR-1 |
LR+/-指的是似然比,LR+越大表示模型对正类的分类越好,LR-越大表示模型对负类的分类效果越好。F1值是精确值和召回率的调和均值,其实原公式是F_{\beta}=(1+\beta^2)\times\frac{\text{P}\times\text{R}}{\beta^2\times\text{P}+\text{R}},这里的\beta表示:召回率的权重是准确率的\beta倍。即F值是一种精确率和召回率的综合指标,权重由\beta决定。MCC值在[-1,1]之间,靠近1表示完全预测正确m靠近-1表示完全悖论,0表示随机预测
def _check_zero_division(zero_division):
if isinstance(zero_division, str) and zero_division == "warn":
return
elif isinstance(zero_division, (int, float)) and zero_division in [0, 1]:
return
raise ValueError('Got zero_division={0}. Must be one of ["warn", 0, 1]'.format(zero_division))
# 权重求和
def _weighted_sum(sample_score, sample_weight, normalize=False):
if normalize:
return np.average(sample_score, weights=sample_weight)
elif sample_weight is not None:
return np.dot(sample_score, sample_weight)
else:
return sample_score.sum()
########################### 计算准确率 #################################
def accuracy_score(y_true, y_pred, *, normalize=True, sample_weight=None):
"""Accuracy classification score.
Parameters
y_true : 1d array-like, or label indicator array / sparse matrix
Ground truth (correct) labels.
y_pred : 1d array-like, or label indicator array / sparse matrix
Predicted labels, as returned by a classifier.
normalize : bool, optional (default=True)
If False, return the number of correctly classified samples.
Otherwise, return the fraction of correctly classified samples.
sample_weight : array-like of shape (n_samples,), default=None
Sample weights.
Returns
score : float
If normalize == True, return the fraction of correctly classified samples (float), else returns the number of correctly classified samples (int).
Examples
>>> from sklearn.metrics import accuracy_score
>>> y_pred = [0, 2, 1, 3]
>>> y_true = [0, 1, 2, 3]
>>> accuracy_score(y_true, y_pred)
0.5
>>> accuracy_score(y_true, y_pred, normalize=False)
2
In the multilabel case with binary label indicators:
>>> import numpy as np
>>> accuracy_score(np.array([[0, 1], [1, 1]]), np.ones((2, 2)))
0.5
"""
# Compute accuracy for each possible representation
y_type, y_true, y_pred = _check_targets(y_true, y_pred) # 检核类型
check_consistent_length(y_true, y_pred, sample_weight) # 检验长度
# 以下为核心代码
if y_type.startswith('multilabel'):
differing_labels = count_nonzero(y_true - y_pred, axis=1)
score = differing_labels == 0
else:
score = y_true == y_pred
return _weighted_sum(score, sample_weight, normalize)
########################### 计算多分类的混淆矩阵 #######################################
def confusion_matrix(y_true, y_pred, *, labels=None, sample_weight=None, normalize=None):
"""Compute confusion matrix to evaluate the accuracy of a classification.
Parameters
y_true : array-like of shape (n_samples,)
Ground truth (correct) target values.
y_pred : array-like of shape (n_samples,)
Estimated targets as returned by a classifier.
labels : array-like of shape (n_classes), default=None
List of labels to index the matrix. This may be used to reorder or select a subset of labels.
sample_weight : array-like of shape (n_samples,), default=None Sample weights.
normalize : {'true', 'pred', 'all'}, default=None
Normalizes confusion matrix over the true (rows), predicted (columns)
conditions or all the population. If None, confusion matrix will not be
normalized.
Returns
C : ndarray of shape (n_classes, n_classes)
Confusion matrix whose i-th row and j-th column entry indicates the number of samples with true label being i-th class and prediced label being j-th class.
Examples
>>> from sklearn.metrics import confusion_matrix
>>> y_true = [2, 0, 2, 2, 0, 1]
>>> y_pred = [0, 0, 2, 2, 0, 2]
>>> confusion_matrix(y_true, y_pred)
array([[2, 0, 0],
[0, 0, 1],
[1, 0, 2]])
>>> y_true = ["cat", "ant", "cat", "cat", "ant", "bird"]
>>> y_pred = ["ant", "ant", "cat", "cat", "ant", "cat"]
>>> confusion_matrix(y_true, y_pred, labels=["ant", "bird", "cat"])
array([[2, 0, 0],
[0, 0, 1],
[1, 0, 2]])
In the binary case, we can extract true positives, etc as follows:
>>> tn, fp, fn, tp = confusion_matrix([0, 1, 0, 1], [1, 1, 1, 0]).ravel()
>>> (tn, fp, fn, tp)
(0, 2, 1, 1)
"""
y_type, y_true, y_pred = _check_targets(y_true, y_pred)
if y_type not in ("binary", "multiclass"):
raise ValueError("%s is not supported" % y_type)
if labels is None:
labels = unique_labels(y_true, y_pred)
else:
labels = np.asarray(labels)
n_labels = labels.size
if n_labels == 0:
raise ValueError("'labels' should contains at least one label.")
elif y_true.size == 0:
return np.zeros((n_labels, n_labels), dtype=np.int)
elif np.all([l not in y_true for l in labels]):
raise ValueError("At least one label specified must be in y_true")
if sample_weight is None:
sample_weight = np.ones(y_true.shape[0], dtype=np.int64)
else:
sample_weight = np.asarray(sample_weight)
check_consistent_length(y_true, y_pred, sample_weight)
if normalize not in ['true', 'pred', 'all', None]:
raise ValueError("normalize must be one of {'true', 'pred', 'all', None}")
n_labels = labels.size
label_to_ind = {y: x for x, y in enumerate(labels)}
# convert yt, yp into index
y_pred = np.array([label_to_ind.get(x, n_labels + 1) for x in y_pred])
y_true = np.array([label_to_ind.get(x, n_labels + 1) for x in y_true])
# intersect y_pred, y_true with labels, eliminate items not in labels
ind = np.logical_and(y_pred < n_labels, y_true < n_labels)
y_pred = y_pred[ind]
y_true = y_true[ind]
# also eliminate weights of eliminated items
sample_weight = sample_weight[ind]
# Choose the accumulator dtype to always have high precision
if sample_weight.dtype.kind in {'i', 'u', 'b'}:
dtype = np.int64
else:
dtype = np.float64
cm = coo_matrix((sample_weight, (y_true, y_pred)),shape=(n_labels, n_labels), dtype=dtype,).toarray() # scipy.sparse.coo_matrix
with np.errstate(all='ignore'):
if normalize == 'true':
cm = cm / cm.sum(axis=1, keepdims=True)
elif normalize == 'pred':
cm = cm / cm.sum(axis=0, keepdims=True)
elif normalize == 'all':
cm = cm / cm.sum()
cm = np.nan_to_num(cm)
return cm
########################### 计算多标签的混淆矩阵 #######################################
def multilabel_confusion_matrix(y_true, y_pred, *, sample_weight=None,
labels=None, samplewise=False):
"""Compute a confusion matrix for each class or sample
Parameters
y_true : 1d array-like, or label indicator array / sparse matrix of shape (n_samples, n_outputs) or (n_samples,)
Ground truth (correct) target values.
y_pred : 1d array-like, or label indicator array / sparse matrix of shape (n_samples, n_outputs) or (n_samples,)
Estimated targets as returned by a classifier
sample_weight : array-like of shape (n_samples,), default=None Sample weights
labels : array-like
A list of classes or column indices to select some (or to force inclusion of classes absent from the data)
samplewise : bool, default=False
In the multilabel case, this calculates a confusion matrix per sample
Returns
multi_confusion : array, shape (n_outputs, 2, 2)
A 2x2 confusion matrix corresponding to each output in the input.
When calculating class-wise multi_confusion (default), then
n_outputs = n_labels; when calculating sample-wise multi_confusion
(samplewise=True), n_outputs = n_samples. If ``labels`` is defined,
the results will be returned in the order specified in ``labels``,
otherwise the results will be returned in sorted order by default.
Multilabel-indicator case:
>>> import numpy as np
>>> from sklearn.metrics import multilabel_confusion_matrix
>>> y_true = np.array([[1, 0, 1],
... [0, 1, 0]])
>>> y_pred = np.array([[1, 0, 0],
... [0, 1, 1]])
>>> multilabel_confusion_matrix(y_true, y_pred)
array([[[1, 0],
[0, 1]],
<BLANKLINE>
[[1, 0],
[0, 1]],
<BLANKLINE>
[[0, 1],
[1, 0]]])
Multiclass case:
>>> y_true = ["cat", "ant", "cat", "cat", "ant", "bird"]
>>> y_pred = ["ant", "ant", "cat", "cat", "ant", "cat"]
>>> multilabel_confusion_matrix(y_true, y_pred,
... labels=["ant", "bird", "cat"])
array([[[3, 1],
[0, 2]],
<BLANKLINE>
[[5, 0],
[1, 0]],
<BLANKLINE>
[[2, 1],
[1, 2]]])
"""
y_type, y_true, y_pred = _check_targets(y_true, y_pred)
if sample_weight is not None:
sample_weight = column_or_1d(sample_weight)
check_consistent_length(y_true, y_pred, sample_weight)
if y_type not in ("binary", "multiclass", "multilabel-indicator"):
raise ValueError("%s is not supported" % y_type)
present_labels = unique_labels(y_true, y_pred)
if labels is None:
labels = present_labels
n_labels = None
else:
n_labels = len(labels)
labels = np.hstack([labels, np.setdiff1d(present_labels, labels,assume_unique=True)])
if y_true.ndim == 1:
if samplewise:
raise ValueError("Samplewise metrics are not available outside of multilabel classification.")
le = LabelEncoder()
le.fit(labels)
y_true = le.transform(y_true)
y_pred = le.transform(y_pred)
sorted_labels = le.classes_
# labels are now from 0 to len(labels) - 1 -> use bincount
tp = y_true == y_pred
tp_bins = y_true[tp]
if sample_weight is not None:
tp_bins_weights = np.asarray(sample_weight)[tp]
else:
tp_bins_weights = None
if len(tp_bins):
tp_sum = np.bincount(tp_bins, weights=tp_bins_weights,
minlength=len(labels))
else:
# Pathological case
true_sum = pred_sum = tp_sum = np.zeros(len(labels))
if len(y_pred):
pred_sum = np.bincount(y_pred, weights=sample_weight,
minlength=len(labels))
if len(y_true):
true_sum = np.bincount(y_true, weights=sample_weight,
minlength=len(labels))
# Retain only selected labels
indices = np.searchsorted(sorted_labels, labels[:n_labels])
tp_sum = tp_sum[indices]
true_sum = true_sum[indices]
pred_sum = pred_sum[indices]
else:
sum_axis = 1 if samplewise else 0
# All labels are index integers for multilabel.
# Select labels:
if not np.array_equal(labels, present_labels):
if np.max(labels) > np.max(present_labels):
raise ValueError('All labels must be in [0, n labels) for '
'multilabel targets. '
'Got %d > %d' %
(np.max(labels), np.max(present_labels)))
if np.min(labels) < 0:
raise ValueError('All labels must be in [0, n labels) for '
'multilabel targets. '
'Got %d < 0' % np.min(labels))
if n_labels is not None:
y_true = y_true[:, labels[:n_labels]]
y_pred = y_pred[:, labels[:n_labels]]
# calculate weighted counts
true_and_pred = y_true.multiply(y_pred)
tp_sum = count_nonzero(true_and_pred, axis=sum_axis,
sample_weight=sample_weight)
pred_sum = count_nonzero(y_pred, axis=sum_axis,
sample_weight=sample_weight)
true_sum = count_nonzero(y_true, axis=sum_axis,
sample_weight=sample_weight)
fp = pred_sum - tp_sum
fn = true_sum - tp_sum
tp = tp_sum
if sample_weight is not None and samplewise:
sample_weight = np.array(sample_weight)
tp = np.array(tp)
fp = np.array(fp)
fn = np.array(fn)
tn = sample_weight * y_true.shape[1] - tp - fp - fn
elif sample_weight is not None:
tn = sum(sample_weight) - tp - fp - fn
elif samplewise:
tn = y_true.shape[1] - tp - fp - fn
else:
tn = y_true.shape[0] - tp - fp - fn
return np.array([tn, fp, fn, tp]).T.reshape(-1, 2, 2)
########################### 计算多标签的精确率/召回率/F分数 #######################################
def precision_recall_fscore_support(y_true, y_pred, *, beta=1.0, labels=None, pos_label=1,
average=None, warn_for=('precision', 'recall', 'f-score'), sample_weight=None,
zero_division="warn"):
"""Compute precision, recall, F-measure and support for each class
Parameters
y_true : 1d array-like, or label indicator array / sparse matrix
Ground truth (correct) target values.
y_pred : 1d array-like, or label indicator array / sparse matrix
Estimated targets as returned by a classifier.
beta : float, 1.0 by default
The strength of recall versus precision in the F-score.
labels : list, optional
The set of labels to include when average != 'binary', and their
order if average is None. Labels present in the data can be
excluded, for example to calculate a multiclass average ignoring a
majority negative class, while labels not present in the data will
result in 0 components in a macro average. For multilabel targets,
labels are column indices. By default, all labels in ``y_true`` and
y_pred are used in sorted order.
pos_label : str or int, 1 by default
The class to report if ``average='binary'`` and the data is binary.
If the data are multiclass or multilabel, this will be ignored;
setting ``labels=[pos_label]`` and ``average != 'binary'`` will report
scores for that label only.
average : string, [None (default), 'binary', 'micro', 'macro', 'samples', 'weighted']
If ``None``, the scores for each class are returned. Otherwise, this
determines the type of averaging performed on the data:
``'binary'``:
Only report results for the class specified by ``pos_label``.
This is applicable only if targets (``y_{true,pred}``) are binary.
``'micro'``:
Calculate metrics globally by counting the total true positives,
false negatives and false positives.
``'macro'``:
Calculate metrics for each label, and find their unweighted
mean. This does not take label imbalance into account.
``'weighted'``:
Calculate metrics for each label, and find their average weighted
by support (the number of true instances for each label). This
alters 'macro' to account for label imbalance; it can result in an
F-score that is not between precision and recall.
``'samples'``:
Calculate metrics for each instance, and find their average (only
meaningful for multilabel classification where this differs from
:func:`accuracy_score`).
warn_for : tuple or set, for internal use
This determines which warnings will be made in the case that this
function is being used to return only one of its metrics.
sample_weight : array-like of shape (n_samples,), default=None
Sample weights.
zero_division : "warn", 0 or 1, default="warn"
Sets the value to return when there is a zero division:
- recall: when there are no positive labels
- precision: when there are no positive predictions
- f-score: both
If set to "warn", this acts as 0, but warnings are also raised.
Returns
precision : float (if average is not None) or array of float, shape =\
[n_unique_labels]
recall : float (if average is not None) or array of float, , shape =\
[n_unique_labels]
fbeta_score : float (if average is not None) or array of float, shape =\
[n_unique_labels]
support : None (if average is not None) or array of int, shape =\
[n_unique_labels]
The number of occurrences of each label in ``y_true``.
Examples
>>> import numpy as np
>>> from sklearn.metrics import precision_recall_fscore_support
>>> y_true = np.array(['cat', 'dog', 'pig', 'cat', 'dog', 'pig'])
>>> y_pred = np.array(['cat', 'pig', 'dog', 'cat', 'cat', 'dog'])
>>> precision_recall_fscore_support(y_true, y_pred, average='macro')
(0.22..., 0.33..., 0.26..., None)
>>> precision_recall_fscore_support(y_true, y_pred, average='micro')
(0.33..., 0.33..., 0.33..., None)
>>> precision_recall_fscore_support(y_true, y_pred, average='weighted')
(0.22..., 0.33..., 0.26..., None)
It is possible to compute per-label precisions, recalls, F1-scores and
supports instead of averaging:
>>> precision_recall_fscore_support(y_true, y_pred, average=None,
... labels=['pig', 'dog', 'cat'])
(array([0. , 0. , 0.66...]),
array([0., 0., 1.]), array([0. , 0. , 0.8]),
array([2, 2, 2]))
"""
_check_zero_division(zero_division)
if beta < 0:
raise ValueError("beta should be >=0 in the F-beta score")
labels = _check_set_wise_labels(y_true, y_pred, average, labels, pos_label)
# Calculate tp_sum, pred_sum, true_sum ###
samplewise = average == 'samples'
MCM = multilabel_confusion_matrix(y_true, y_pred, sample_weight=sample_weight,
labels=labels, samplewise=samplewise)
tp_sum = MCM[:, 1, 1]
pred_sum = tp_sum + MCM[:, 0, 1]
true_sum = tp_sum + MCM[:, 1, 0]
if average == 'micro':
tp_sum = np.array([tp_sum.sum()])
pred_sum = np.array([pred_sum.sum()])
true_sum = np.array([true_sum.sum()])
# Finally, we have all our sufficient statistics. Divide! #
beta2 = beta ** 2
# Divide, and on zero-division, set scores and/or warn according to
# zero_division:
precision = _prf_divide(tp_sum, pred_sum, 'precision','predicted',
average, warn_for, zero_division)
recall = _prf_divide(tp_sum, true_sum, 'recall', 'true',
average, warn_for, zero_division)
# warn for f-score only if zero_division is warn, it is in warn_for
# and BOTH prec and rec are ill-defined
if zero_division == "warn" and ("f-score",) == warn_for:
if (pred_sum[true_sum == 0] == 0).any():
_warn_prf(average, "true nor predicted", 'F-score is', len(true_sum))
# if tp == 0 F will be 1 only if all predictions are zero, all labels are
# zero, and zero_division=1. In all other case, 0
if np.isposinf(beta):
f_score = recall
else:
denom = beta2 * precision + recall
denom[denom == 0.] = 1 # avoid division by 0
f_score = (1 + beta2) * precision * recall / denom
# Average the results
if average == 'weighted':
weights = true_sum
if weights.sum() == 0:
zero_division_value = 0.0 if zero_division in ["warn", 0] else 1.0
# precision is zero_division if there are no positive predictions
# recall is zero_division if there are no positive labels
# fscore is zero_division if all labels AND predictions are
# negative
return (zero_division_value if pred_sum.sum() == 0 else 0,
zero_division_value,
zero_division_value if pred_sum.sum() == 0 else 0,
None)
elif average == 'samples':
weights = sample_weight
else:
weights = None
if average is not None:
assert average != 'binary' or len(precision) == 1
precision = np.average(precision, weights=weights)
recall = np.average(recall, weights=weights)
f_score = np.average(f_score, weights=weights)
true_sum = None # return no support
return precision, recall, f_score, true_sum
- AP与mAP
https://www.jianshu.com/p/ba1f7895b429
二分类问题的P-R曲线(precision-recall curve),P-R曲线下面与x轴围成的面积称为average precision(AP)。显然通过积分来计算
但通常情况下都是使用估算或者插值的方式计算。
mAP(mean average precision)的意义是为了评估你整个目标检测模型的准确度。方法是:计算每个分类的AP,求和再平均,得到的就是mAP
- F_\beta Score
Precision和Recall是互相影响的,理想情况下肯定是做到两者都高,但是一般情况下Precision高、Recall就低,Recall高、Precision就低。为了均衡两个指标,我们可以采用Precision和Recall的加权调和平均(weighted harmonic mean)来衡量,即F_\beta Score,公式如下:
\beta表示权重,
当\beta \rightarrow 0 : F_{\beta} \approx P;当\beta \rightarrow \infty:F_{\beta} \approx R。通俗的语言就是:\beta越大,Recall的权重越大,\beta越小,Precision的权重越大。
随着如\beta=1为F_\beta此时Precision和Recall的权重相等,公式如下:
由于F_\beta Score无法直观反映数据的情况,同时业务含义相对较弱,实际工作用到的不多。
- ROC和AUC
AUC是一种模型分类指标,且仅仅是二分类模型的评价指标。AUC是Area Under Curve的简称,那么Curve就是ROC(Receiver Operating Characteristic),翻译为"接受者操作特性曲线"。也就是说ROC是一条曲线,AUC是一个面积值。
ROC:ROC曲线为FPR与TPR之间的关系曲线,这个组合以 FPR 对TPR,即是以代价(costs)对收益(benefits),显然收益越高,代价越低,模型的性能就越好。
x轴为假阳性率(FPR):在所有的负样本中,分类器预测错误的比例
y轴为真阳性率(TPR):在所有的正样本中,分类器预测正确的比例(等于Recall)
为了更好地理解ROC曲线,我们使用具体的实例来说明:
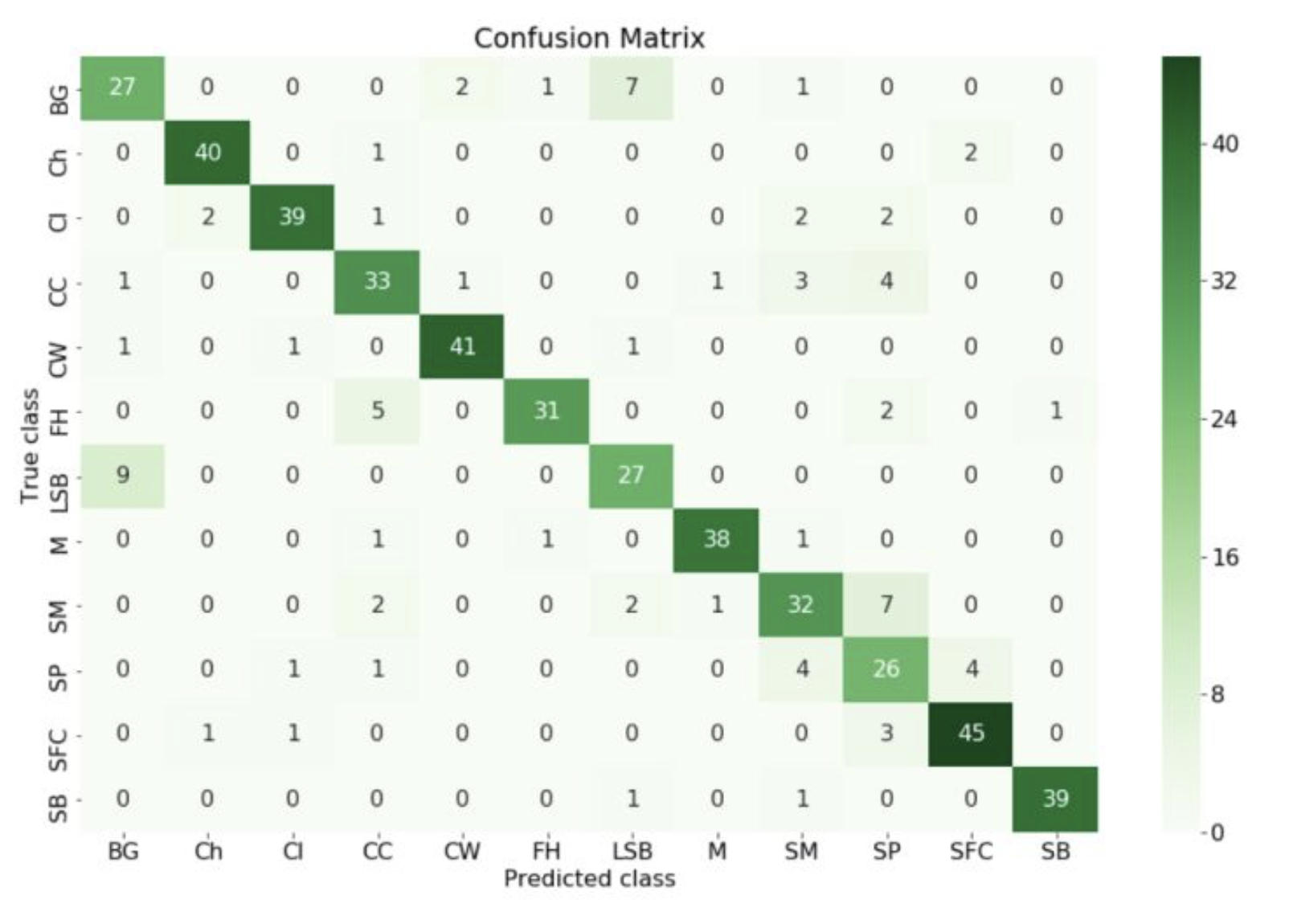
如在医学诊断的主要任务是尽量把生病的人群都找出来,也就是TPR越高越好。而尽量降低没病误诊为有病的人数,也就是FPR越低越好。不难发现,这两个指标之间是相互制约的。如果某个医生对于有病的症状比较敏感,稍微的小症状都判断为有病,那么他的TPR应该会很高,但是FPR也就相应地变高。最极端的情况下,他把所有的样本都看做有病,那么TPR达到1,FPR也为1。以FPR为横轴,TPR为纵轴,得到如下ROC空间。
可以看出,左上角的点(TPR=1,FPR=0)为完美分类,也就是这个医生医术高明,诊断全对。点A(TPR>FPR),医生A的判断大体是正确的。中线上的点B(TPR=FPR),也就是医生B全都是蒙的,蒙对一半,蒙错一半;下半平面的点C(TPR<FPR),这个医生说你有病,那么你很可能没有病,医生C的话我们要反着听,为真庸医。上图中一个阈值,得到一个点。现在我们需要一个独立于阈值的评价指标来衡量这个医生的医术如何,也就是遍历所有的阈值,得到ROC曲线。
假设下图是某医生的诊断统计图,为未得病人群(上图)和得病人群(下图)的模型输出概率分布图(横坐标表示模型输出概率,纵坐标表示概率对应的人群的数量),显然未得病人群的概率值普遍低于得病人群的输出概率值(即正常人诊断出疾病的概率小于得病人群诊断出疾病的概率)。

竖线代表阈值。显然,图中给出了某个阈值对应的混淆矩阵,通过改变不同的阈值1.0 \rightarrow 0,得到一系列的混淆矩阵,进而得到一系列的TPR和FPR,绘制出ROC曲线。
阈值为1时,不管你什么症状,医生均未诊断出疾病(预测值都为N,此时绿色和红色区域的面积为0,因此FPR=TPR=0,位于左下。随着阈值的减小,红色和绿色区域增大,紫色和蓝色区域减小。阈值为0时,不管你什么症状,医生都诊断结果都是得病(预测值都为P),此时绿色和红色区域均占整个区域,即紫色和蓝色区域的面积为0,此时 FPR=TPR=1,位于右上。
AUC:AUC值为ROC曲线所覆盖的区域面积,显然,AUC越大,分类器分类效果越好。
AUC=1,是完美分类器。
0.5<AUC<1,优于随机猜测,有预测价值。
AUC=0.5,跟随机猜测一样(例:丢铜板),没有预测价值。
AUC < 0.5,比随机猜测还差;但只要总是反预测而行,就优于随机猜测。
注: 对于AUC小于0.5的模型,我们可以考虑取反(模型预测为positive,那我们就取negtive),这样就可以保证模型的性能不可能比随机猜测差。
以下为ROC曲线和AUC值的实例:
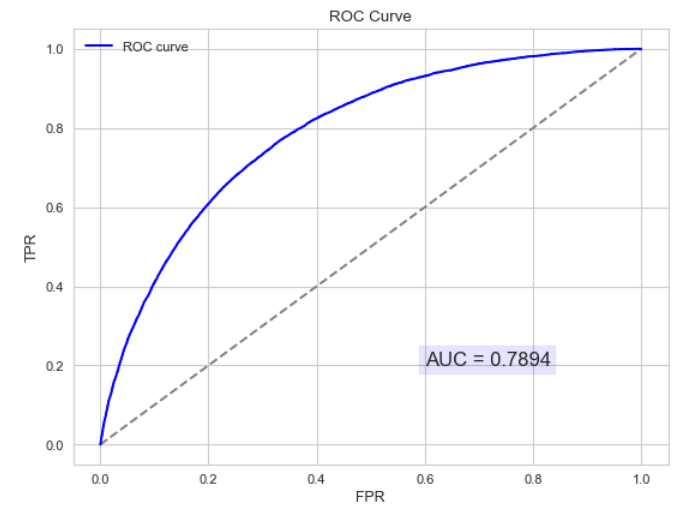
AUC的物理意义:正样本的预测结果大于负样本的预测结果的概率。所以AUC反应的是分类器对样本的排序能力。另外值得注意的是,AUC对样本类别是否均衡并不敏感,这也是不均衡样本通常用AUC评价分类器性能的一个原因。AUC只关注正负样本之间的排序,并不关心正样本内部,或者负样本内部的排序。这也体现了AUC的本质:任意个正样本的概率都大于负样本的概率的能力
AUC的计算:
(1). AUC为ROC曲线下的面积,那我们直接计算面积可得。面积为一个个小的梯形面积(曲线)之和。计算的精度与阈值的精度有关。
(2). 根据AUC的物理意义,我们计算正样本预测结果大于负样本预测结果的概率。取n_1*n_0(n_1为正样本数,n_0为负样本数)个二元组,比较score(预测结果),最后得到AUC。时间复杂度为O(N*M)。
(3). 首先把所有样本按照score排序,依次用rank表示他们,如最大score的样本,rank=n(n=n_0+n_1,其中n_0为负样本个数,n_1为正样本个数),其次为n-1。对于正样本中rank最大的样本rank\_max,有n_1-1个其他正样本比它小。有rank\_max-n_1个负样本比它小。其次为rank\_second-(n_1-1)。最后我们得到正样本大于负样本的概率为:
AUC=\frac{\sum_{\text{正样本}}rank(core)-n_1*(n_1+2)/2}{n_0*n_1}(4). 计算复杂度为O(N+M)。
ROC和AUC都能应用于非均衡的分类问题:ROC曲线只与横坐标(FPR)和纵坐标(TPR)有关系,其中:
以及混淆矩阵:
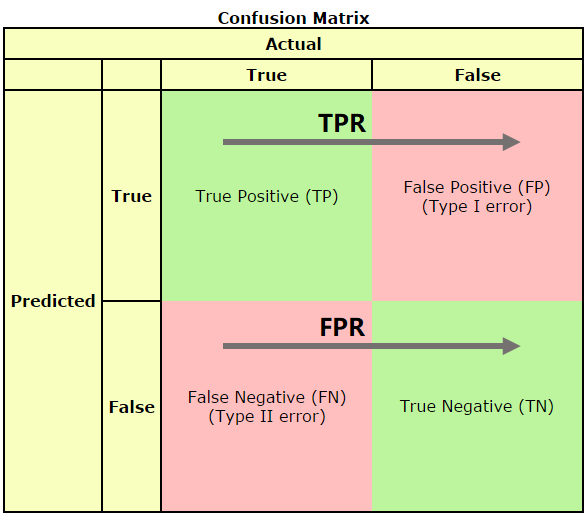
可以发现TPR只是正样本中(第一行)预测正确的概率,在正样本内部进行,并没有牵扯到负样本。而FPR只是负样本中(第二行)预测错误的概率,在负样本内部进行,并没有牵扯到正样本。TPR和FPR的计算并没有涉及正负样本的互动(也就是没有跨行)。和正负样本的比例没有关系。因此ROC的值与实际的正负样本比例无关,因此既可以用于均衡问题,也可以用于非均衡问题。而AUC的几何意义为ROC曲线下的面积,因此也和实际的正负样本比例无关。
- KS(Kolmogorov-Smirnov)
KS值是在模型中用于区分预测正负样本分隔程度的评价指标,一般应用于金融风控领域。与ROC曲线相似,ROC是以FPR作为横坐标,TPR作为纵坐标,通过改变不同阈值,从而得到ROC曲线。而在KS曲线中,则是以阈值作为横坐标,以FPR和TPR作为纵坐标,ks曲线则为TPR-FPR,ks曲线的最大值通常为ks值。
为什么这样求KS值呢?我们知道,当阈值减小时,TPR和FPR会同时减小,当阈值增大时,TPR和FPR会同时增大。而在实际工程中,我们希望TPR更大一些,FPR更小一些,即TPR-FPR越大越好,即ks值越大越好。KS值的取值范围是[0,1]。通常来说,值越大,模型区分正负样本的能力越强(一般0.3以上,说明模型的效果比较好)。
以下为KS曲线的实例 :

- micro与macro
假如我们有n个二分类混淆矩阵,评价模型通常有两种方式一种叫macro,一种叫micro。
macro方法
计算出各混淆矩阵的Recall,Precision,记为(P_1,R_1),(P_2,R_2),\cdots,(P_n,R_n):
对各个混淆矩阵的Recall,Precision求平均,然后再根据求得的Recall,Precision计算F1。
micro方法
将各混淆矩阵对应的元素进行平均,得到平均混淆矩阵:
再基于平均混淆矩阵计算Recall,Precision,然后再根据求得的Recall,Precision计算F1:
1.4 聚类指标
- 兰德指数
- 轮廓系数
- 互信息
1.5 检测指标
- IOU
https://zhuanlan.zhihu.com/p/112057799
IOU定义了两个bounding box的重叠度,可以说,当算法给出的框(bounding box)和人工标注的框(ground truth)差异很小时,即重叠度很大时,那么算法产生的boundingbox就很准确。
IOU正是表达这种bounding box和ground truth的差异的指标。
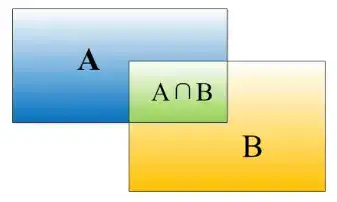
如上图,矩形框A、B的一个重合度IOU计算公式为(交集除以并集): IoU=(A\cap B)/(A\cup B)
def bb_intersection_over_union(boxA, boxB):
boxA = [int(x) for x in boxA]
boxB = [int(x) for x in boxB]
xA = max(boxA[0], boxB[0])
yA = max(boxA[1], boxB[1])
xB = min(boxA[2], boxB[2])
yB = min(boxA[3], boxB[3])
interArea = max(0, xB - xA + 1) * max(0, yB - yA + 1)
boxAArea = (boxA[2] - boxA[0] + 1) * (boxA[3] - boxA[1] + 1)
boxBArea = (boxB[2] - boxB[0] + 1) * (boxB[3] - boxB[1] + 1)
iou = interArea / float(boxAArea + boxBArea - interArea)
return iou
- GIou
参考论文:Generalized Intersection over Union: A Metric and A Loss for Bounding Box Regression
IoU是检测任务中最常用的指标,由于IoU是比值的概念,对目标物体的scale是不敏感的。然而检测任务中的BBox的回归损失(MSE loss, l1-smooth loss等)优化和IoU优化不是完全等价的(见下图)。而且L_n范数对物体的scale也比较敏感。这篇论文提出可以直接把IoU设为回归的loss。然而有个问题是IoU无法直接优化没有重叠的部分,为了解决这个问题这篇paper提出了GIoU的思想。
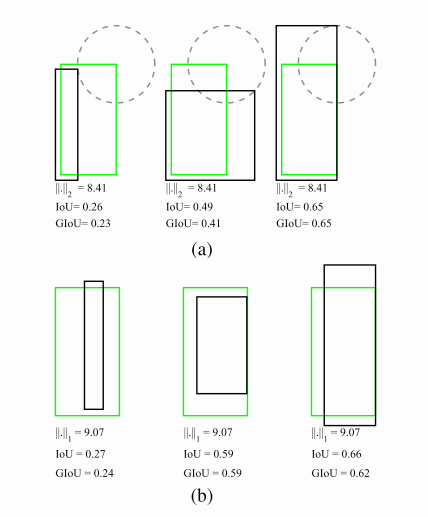
要将IoU设计为损失,主要需要解决两个问题:
预测值和Ground truth没有重叠的话,IoU始终为0且无法优化
IoU无法辨别不同方式的对齐,比如方向不一致等。
假如现在有两个任意性质A,B,我们找到一个最小的封闭形状C,让C可以把A,B包含在内,然后我们计算C中没有覆盖A和B的面积占C总面积的比值,然后用A与B的IoU减去这个比值:
GIoU有如下性质:
与IoU类似,GIoU也可以作为一个距离,loss可以用 [公式] 来计算 同原始IoU类似,GIoU对物体的大小不敏感 GIoU总是小于等于IoU,对于IoU,有 [公式] ,GIoU则是 [公式] 。在两个形状完全重合时,有 [公式] 由于GIoU引入了包含A,B两个形状的C,所以当A,B不重合时,依然可以进行优化。
总之就是保留了IoU的原始性质同时弱化了它的缺点,于是论文认为可以将其作为IoU的替代。
我们以2D detecation计算GIoU损失为例:
假设我们现在有预测的Bbox和groud truth的Bbox的坐标,分别记为: B^p=\{x_1^p,y_1^p,x_2^p,y_2^p\},B^g=\{x_1^g,y_1^g,x_2^g,y_2^g\}。注意我们规定对于预测的BBox来说,有x_2^p>x_1^p,y_2^p>y_1^p,主要是为了方便之后点的对应关系。
- 计算B^g的面积:A^g=(x_2^g-x_1^g)*(y_2^g-y_1^g)
- 计算B^p的面积:A^p=(x_2^p-x_1^p)*(y_2^p-y_1^p)
- 计算B^g,B^p的重叠面积:
x_1^I = max(\hat x_1^p, x_1^g), x_2^I = min(\hat x_2^p, x_2^g) \\ y_1^I = max(\hat y_1^p,y_1^g), x_2^I = min(\hat y_2^p, y_2^g) \\ I = \begin{equation} \left\{ \begin{array}{**lr**} (x_2^I-x_1^I)*(y_2^I-y_1^I) &x_2^I>x_1^I, y_2^I>y_1^I\\ 0 &otherwise& \end{array} \right. \end{equation}
- 找到可以包含B^p,B^g的最小boxB^c
x_1^c = min(\hat x_1^p, x_1^g), x_2^c = max(\hat x_2^p, x_2^g)\\ y_1^c = min(\hat y_1^p,y_1^g), x_2^c = max(\hat y_2^p, y_2^g)
计算B^c的面积: A^c = (x_2^c-x_1^c) *(y_2^c-y_1^c)
计算IoU:IoU = \frac{I}{U}=\frac{I}{A^p+A^g - I}
计算GIoU = IoU - \frac{A^c - U}{A^c}
8.计算最终的损失:L_{GIoU} = 1 - GIoU
def Giou(rec1,rec2):
x1,x2,y1,y2 = rec1 #分别是第一个矩形左右上下的坐标
x3,x4,y3,y4 = rec2
iou = Iou(rec1,rec2)
area_C = (max(x1,x2,x3,x4)-min(x1,x2,x3,x4))*(max(y1,y2,y3,y4)-min(y1,y2,y3,y4))
area_1 = (x2-x1)*(y1-y2)
area_2 = (x4-x3)*(y3-y4)
sum_area = area_1 + area_2
w1 = x2 - x1#第一个矩形的宽
w2 = x4 - x3#第二个矩形的宽
h1 = y1 - y2
h2 = y3 - y4
W = min(x1,x2,x3,x4)+w1+w2-max(x1,x2,x3,x4)#交叉部分的宽
H = min(y1,y2,y3,y4)+h1+h2-max(y1,y2,y3,y4)#交叉部分的高
Area = W*H#交叉的面积
add_area = sum_area - Area #两矩形并集的面积
end_area = (area_C - add_area)/area_C #(c/(AUB))/c的面积
giou = iou - end_area
return giou
- NMS
https://zhuanlan.zhihu.com/p/60794316
非极大值抑制(non maximum suppression,nms)是通常用于目标检测算法,作用是去除重复的区域,就是抑制不是极大值的元素,在这里就是去除和想要的框重叠部分过大的框。
NMS的基本思想是将所有框按得分进行排序,然后无条件保留其中得分最高的框,然后遍历其余框找到和当前最高分的框的重叠面积(IOU)大于一定阈值的框,并删除。然后继续这个过程,找另一个得分高的框,再删除和其IOU大于阈值的框,一直循环直到所有的框都被处理。
在目标检测中,常用非极大值抑制算法(NMS)对生成的大量候选框进行后处理,在faster R-CNN中,每一个bbox都有一个得分,然后使用NMS去除冗余的候选框,得到最具代表性的bbox以加快目标检测的效率。

NMS的具体实现流程为:
- 根据候选框的类别分类概率(得分),按最高到最低将BBox排序,例如:A>B>C>D>E>F
- 先标记最大概率矩形框A是要保留下来的,即A的分数最高,则无条件保留
- 将B~E分别与A求重叠率IoU(两框的交并比),假设B、D与A的IoU大于设定的阈值,那么B和D可以认为是重复标记被剔除
- 继续从剩下的矩形框C、E、F中选择概率最大的C,标记为要无条件保留下来的框,然后分别计算C与E、F的重叠度,扔掉重叠度超过设定阈值的矩形框
- 就这样一直重复下去,直到剩下的矩形框没有了,得到所有要保留下来的矩形框
import numpy as np
def non_max_suppression_fast(boxes, overlapThresh):
"""
boxes: boxes为一个m*n的矩阵,m为bbox的个数,n的前4列为每个bbox的坐标,
格式为(x1,y1,x2,y2),有时会有第5列,该列为每一类的置信
overlapThresh: 最大允许重叠率
"""
# if there are no boxes, return an empty list
if len(boxes) == 0:
return []
# if the bounding boxes are integers, convert them to floats
# this is important since we'll be doing a bunch of divisions
if boxes.dtype.kind == "i":
boxes = boxes.astype("float")
# initialize the list of picked indexes
pick = []
# grab the coordinates of all bounding boxes respectively
x1 = boxes[:,0] # startX
y1 = boxes[:,1] # startY
x2 = boxes[:,2] # endX
y2 = boxes[:,3] # endY
# probs = boxes[:,4]
# compute the area of the bounding boxes and sort the bboxes
# by the bottom y-coordinate of the bboxes by ascending order
# and grab the indexes of the sorted coordinates of bboxes
area = (x2 - x1 + 1) * (y2 - y1 + 1)
idxs = np.argsort(y2)
# if probabilities are provided, sort by them instead
# idxs = np.argsort(probs)
# keep looping while some indexes still remain in the idxs list
while len(idxs) > 0:
# grab the last index in the idxs list (the bottom-right box)
# and add the index value to the list of picked indexes
last = len(idxs) - 1
i = idxs[last]
pick.append(i)
# find the largest coordinates for the start of the bbox
# and the smallest coordinates for the end of the bbox
# in the rest of bounding boxes.
xx1 = np.maximum(x1[i], x1[idxs[:last]])
yy1 = np.maximum(y1[i], y1[idxs[:last]])
xx2 = np.minimum(x2[i], x2[idxs[:last]])
yy2 = np.minimum(y2[i], y2[idxs[:last]])
# the width and height of the bounding box
w = np.maximum(0, xx2 - xx1 + 1)
h = np.maximum(0, yy2 - yy1 + 1)
# the ratio of overlap in the bounding box
overlap = (w * h) / area[idxs[:last]]
# delete all indexes from the index list that overlap is larger than overlapThresh
idxs = np.delete(idxs, np.concatenate(([last], np.where(overlap > overlapThresh)[0])))
# return only the bounding boxes that were picked using the
# integer data type
return boxes[pick].astype("int")
- soft_nms
这是对nms的改进,它没有删除所有IOU大于阈值的框,而是降低其置信度。
def cpu_soft_nms(np.ndarray[float, ndim=2] boxes, float sigma=0.5, float Nt=0.3, float threshold=0.001, unsigned int method=0):
cdef unsigned int N = boxes.shape[0]
cdef float iw, ih, box_area
cdef float ua
cdef int pos = 0
cdef float maxscore = 0
cdef int maxpos = 0
cdef float x1,x2,y1,y2,tx1,tx2,ty1,ty2,ts,area,weight,ov
for i in range(N): 每次找最大的得分和相应的box
maxscore = boxes[i, 4]
maxpos = i
tx1 = boxes[i,0]
ty1 = boxes[i,1]
tx2 = boxes[i,2]
ty2 = boxes[i,3]
ts = boxes[i,4]
pos = i + 1
# get max box
while pos < N:
if maxscore < boxes[pos, 4]:
maxscore = boxes[pos, 4]
maxpos = pos
pos = pos + 1
# add max box as a detection
boxes[i,0] = boxes[maxpos,0]
boxes[i,1] = boxes[maxpos,1]
boxes[i,2] = boxes[maxpos,2]
boxes[i,3] = boxes[maxpos,3]
boxes[i,4] = boxes[maxpos,4]
# swap ith box with position of max box 把得分最大的放到当前第一个位置
boxes[maxpos,0] = tx1
boxes[maxpos,1] = ty1
boxes[maxpos,2] = tx2
boxes[maxpos,3] = ty2
boxes[maxpos,4] = ts
tx1 = boxes[i,0]
ty1 = boxes[i,1]
tx2 = boxes[i,2]
ty2 = boxes[i,3]
ts = boxes[i,4]
pos = i + 1
# NMS iterations, note that N changes if detection boxes fall below threshold
while pos < N: 当前第一个,也就是得分最高的一个,和后面所有的box进行nms操作
x1 = boxes[pos, 0]
y1 = boxes[pos, 1]
x2 = boxes[pos, 2]
y2 = boxes[pos, 3]
s = boxes[pos, 4]
area = (x2 - x1 + 1) * (y2 - y1 + 1)
iw = (min(tx2, x2) - max(tx1, x1) + 1) width的重叠部分长度
if iw > 0:
ih = (min(ty2, y2) - max(ty1, y1) + 1) height的重叠部分长度
if ih > 0:
ua = float((tx2 - tx1 + 1) * (ty2 - ty1 + 1) + area - iw * ih)
ov = iw * ih / ua #iou between max box and detection box
if method == 1: # linear
if ov > Nt:
weight = 1 - ov
else:
weight = 1
elif method == 2: # gaussian
weight = np.exp(-(ov * ov)/sigma)
else: # original NMS
if ov > Nt:
weight = 0
else:
weight = 1
boxes[pos, 4] = weight*boxes[pos, 4]
# if box score falls below threshold, discard the box by swapping with last box
# update N
if boxes[pos, 4] < threshold:
boxes[pos,0] = boxes[N-1, 0]
boxes[pos,1] = boxes[N-1, 1]
boxes[pos,2] = boxes[N-1, 2]
boxes[pos,3] = boxes[N-1, 3]
boxes[pos,4] = boxes[N-1, 4]
N = N - 1
pos = pos - 1
pos = pos + 1
keep = [i for i in range(N)]
return keep
- MSER
MSER全称叫做最大稳定极值区域(MSER-Maximally Stable Extremal Regions),是一种检测图像中文本区域的传统图像算法,主要是基于分水岭的思想来对图像进行斑点(blob)区域检测。
就准确率来说,MSER对文本区域的检测效果是不能和深度学习如CTPN、Pixellink等相比的,但是可以用于自然场景的文本检测的前期阶段,产生尽可能多的proposals。
MSER对灰度图像取阈值进行二值化处理,阈值从0到255依次进行递增,阈值的递增类似于分水岭算法中的水平面的上升,随着水平面的上升,有一些山谷和较矮的丘陵会被淹没,如果从天空往下看,则整个区域被分为陆地和水域两个部分,这类似于二值图像。图像中灰度值的不同就对应地势高低的不同,每个阈值都都会生成一个二值图。
随着阈值的增加,首先会看到一个全白图像,然后出现小黑点,随着阈值的增加,黑色部分会逐渐增大, 这些黑色区域最终会融合,直到整个图像变成黑色。在得到的所有二值图像中,图像中的某些连通区域变化很小,甚至没有变化,则该区域就被称为最大稳定极值区域。

在一幅含有文字的图像上,由于文字区域的灰度值是一致的,而且和文字周边像素的灰度值差别较大,因此在水平面(阈值)持续增长的一段时间内它们都不会被覆盖,直到阈值涨到文字本身的灰度值时才会被淹没,所以文字区域可以作为最大稳定极值区域。
所以如果一个区域在给定的阈值范围内保持其形状和大小基本稳定不变,而不会与其他区域合并,该区域被认为是稳定的。
上述做法只能检测出灰度图像的黑色区域,不能检测出白色区域,因此还需要对原图进行反转(负片图像),然后再进行阈值从0~255递增的二值化处理过程。这两种操作分别被称为MSER+和MSER-。
MSER 的公式如下:
R(i)表示阈值为i时的某一连通区域,Δ为灰度阈值的微小增加量,q(i)为阈值是i时的区域R(i)的变化率,|R(i)|表示区域R(i)的面积。当q(i)为局部极小值时,说明R(i)区域的变化非常小,则R(i)为可以被认为是最大稳定极值区域。
该算法可以用来粗略地寻找图像中的文字区域,虽然算法思想简单,但要做到效果又快又好还是需要一定基础的,OpenCV直接提供了该算法的接口。
'''
MSER+NMS实现图像文本区域检测
'''
path = '/home/zxd/Downloads/co120110121029-12.jpg'
img = cv2.imread(path)
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY) # 灰度图
vis_1 = img.copy()
vis_2 = img.copy()
# get mser object
mser = cv2.MSER_create(_delta=5, _min_area=10, _max_variation=0.5)
# Detect MSER regions
regions, boxes = mser.detectRegions(gray)
# 绘制文本区域(不规则轮廓)
hulls = [cv2.convexHull(p.reshape(-1, 1, 2)) for p in regions]
cv2.polylines(img, hulls, 1, (0, 255, 0), 1)
# 两种绘制矩形轮廓
# for box in boxes:
# x, y, w, h = box
# cv2.rectangle(vis_1, (x,y),(x+w, y+h), (255, 0, 0), 1)
keep = []
for hull in hulls:
x, y, w, h = cv2.boundingRect(hull)
keep.append([x, y, x + w, y + h])
cv2.rectangle(vis_1, (x, y), (x + w, y + h), (255, 0, 0), 1)
print("%d bounding boxes before nms" % (len(keep)))
# 使用非极大值抑制获取不重复的矩形框
pick = non_max_suppression_fast(np.array(keep), overlapThresh=0.4)
print("%d bounding boxes after nms" % (len(pick)))
# loop over the picked bounding boxes and draw them
for (startX, startY, endX, endY) in pick:
cv2.rectangle(vis_2, (startX, startY), (endX, endY), (0, 0, 255), 1)
# 合并图片
boxing_list = [img, vis_1, vis_2]
boxing = np.concatenate(boxing_list, axis=0)
plt.figure(figsize=(10, 20)) # w,h
plt.imshow(boxing, cmap='gray')
plt.show()
text_mask = np.zeros((img.shape[0], img.shape[1]), dtype=np.uint8)
for contour in hulls:
cv2.drawContours(text_mask, [contour], -1, (255, 255, 255), -1)
img = cv2.imread(path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
text_region = cv2.bitwise_and(img, img, mask=mask)
二、基本结构
2.1 卷积概念
从数学上讲,卷积就是一种运算。某种运算,能被定义出来,至少有以下特征:
首先是抽象的、符号化的
其次,在生活、科研中,有着广泛的作用
比如加法:a+b是抽象的,本身只是一个数学符号。在现实中,有非常多的意义,比如增加、合成、旋转等等。
- 卷积的定义
我们称(f*g)(n)为f,g的卷积,其连续的定义为:
其离散的定义为:
这两个式子有一个共同的特征:

这个特征有什么意义?我们令x=\tau,y=n-\tau,那么x+y=n就是下面这些直线:

如果遍历这些直线,就好比,把毛巾沿着角卷起来:
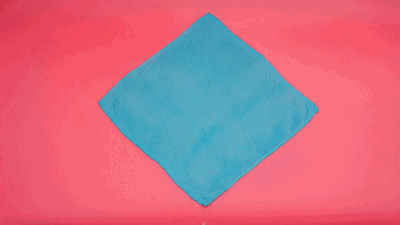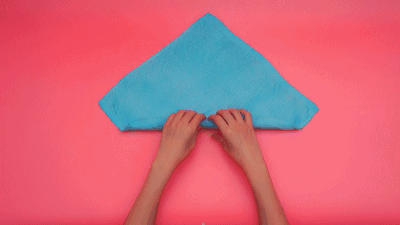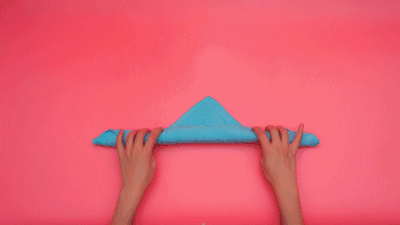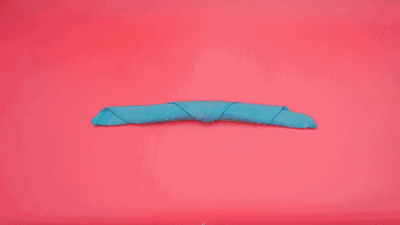
或许,这就是"卷积"名字的来源吧。
- 离散卷积的例子:丢骰子
我有两枚骰子:

把这两枚骰子都抛出去:

求:

这里问题的关键是,两个骰子加起来要等于4,这正是卷积的应用场景。我们把骰子各个点数出现的概率表示出来:

那么,两枚骰子点数加起来为4的情况有:
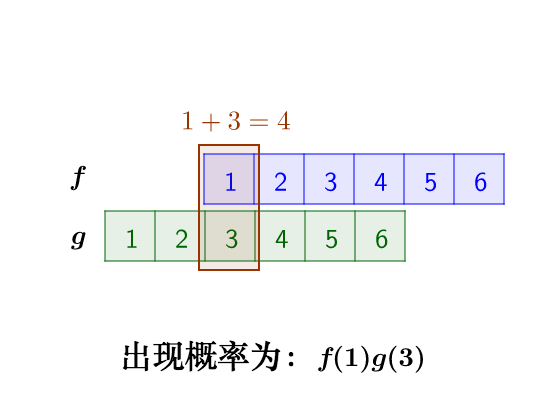
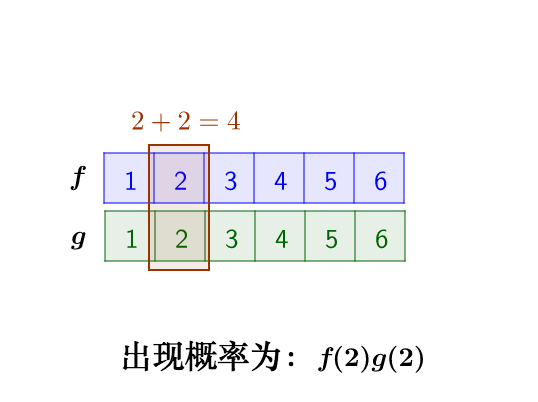

因此,两枚骰子点数加起来为4的概率为:f(1)g(3)+f(2)g(2)+f(3)g(1)
符合卷积的定义,把它写成标准的形式就是:
- 连续卷积的例子:做馒头
楼下早点铺子生意太好了,供不应求,就买了一台机器,不断的生产馒头。假设馒头的生产速度是f(t),那么一天后生产出来的馒头总量为:
馒头生产出来之后,就会慢慢腐败,假设腐败函数为g(t),比如,10个馒头,24小时会腐败:10*g(t)
想想就知道,第一个小时生产出来的馒头,一天后会经历24小时的腐败,第二个小时生产出来的馒头,一天后会经历23小时的腐败。如此,我们可以知道,一天后,馒头总共腐败了:
这就是连续的卷积。
- 图像处理中的应用
原理:有这么一副图像,可以看到,图像上有很多噪点:

高频信号,就好像平地耸立的山峰:
<img src="http://pic1.zhimg.com/80/v2-294698966c5a833cd750df70c0a00c21_hd.jpg)
看起来很显眼。平滑这座山峰的办法之一就是,把山峰刨掉一些土,填到山峰周围去。用数学的话来说,就是把山峰周围的高度平均一下。平滑后得到:

计算:卷积可以帮助实现这个平滑算法。有噪点的原图,可以把它转为一个矩阵:

然后用下面这个平均矩阵(说明下,原图的处理实际上用的是正态分布矩阵,这里为了简单,就用了算术平均矩阵)来平滑图像:
记得刚才说过的算法,把高频信号与周围的数值平均一下就可以平滑山峰。比如我要平滑a_{1,1}4点,就在矩阵中,取出a_{1,1}点附近的点组成矩阵f和g进行卷积计算后,再填回去:
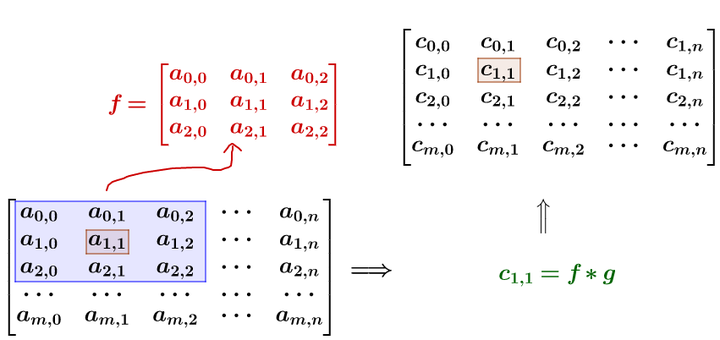
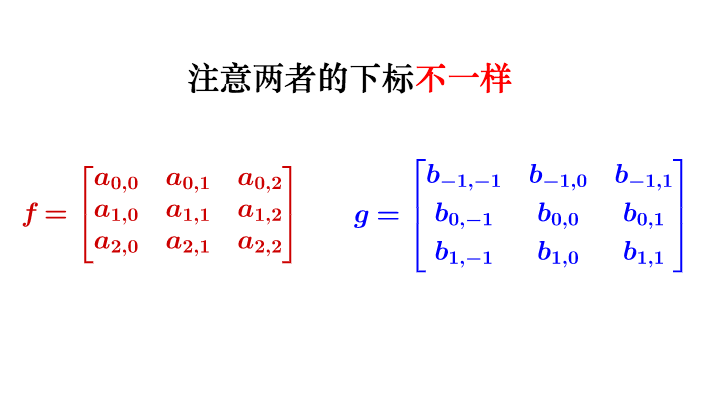
要注意一点,为了运用卷积,g虽然和4f$同维度,但下标有点不一样:
我用一个动图来说明下计算过程:

写成卷积公式就是:
要求c_{4,5},一样可以套用上面的卷积公式。这样相当于实现了g这个矩阵在原来图像上的划动(准确来说,下面这幅图把g矩阵旋转了180^\circ):
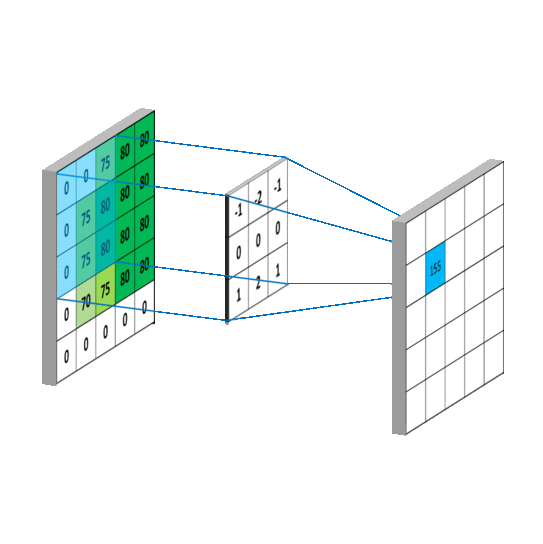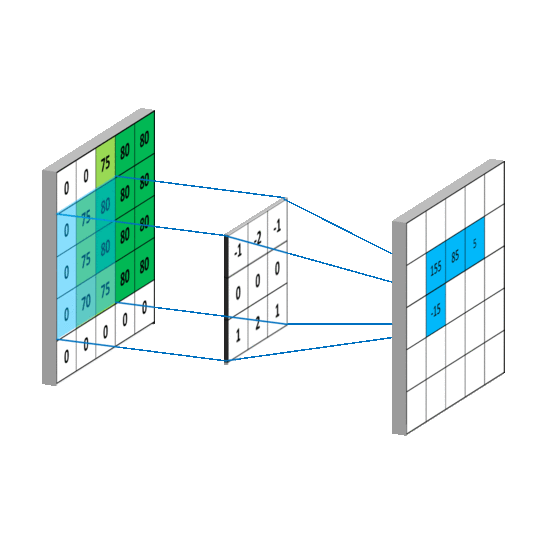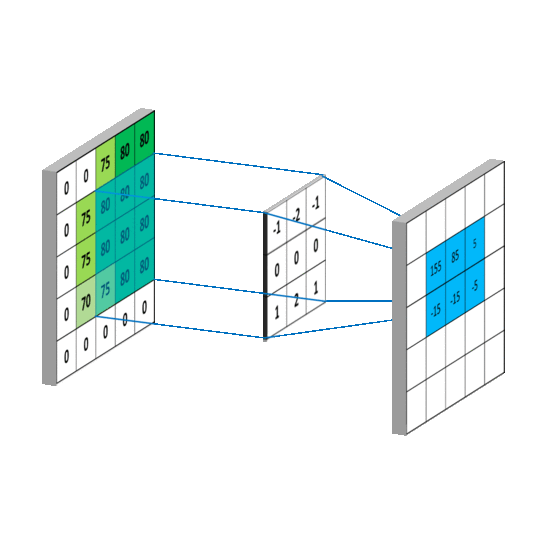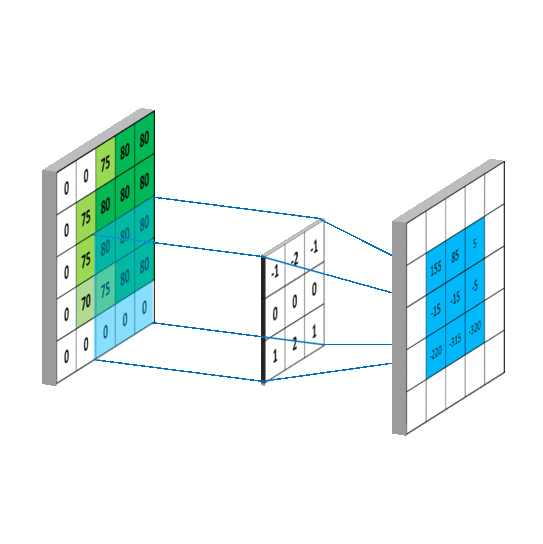
2.2 卷积类型
2.2.1 普通卷积
首先,定义下卷积层的结构参数。


卷积核为3、步幅为1和带有边界扩充的二维卷积结构
卷积核大小(Kernel Size):定义了卷积操作的感受野,在二维卷积中,通常设置为3,即卷积核大小为
3×3。步幅(Stride):定义了卷积核遍历图像时的步幅大小,其默认值通常设置为1,也可将步幅设置为2后对图像进行下采样,这种方式与最大池化类似。
边界扩充(Padding):定义了网络层处理样本边界的方式,当卷积核大于1且不进行边界扩充,输出尺寸将相应缩小;当卷积核以标准方式进行边界扩充,则输出数据的空间尺寸将与输入相等。
输入与输出通道(Channels)构建卷积层时需定义输入通道I,并由此确定输出通道O,这样,可算出每个网络层的参数量为
I×O×K,其中K为卷积核的参数个数。例某个网络层有64个大小为3×3的卷积核,则对应K值为3×3=9。
输出特征的大小:
2.2.2 空洞卷积
参考论文: Multi-Scale Context Aggregation by Dilated Convolutions
空洞卷积(atrous convolutions)又名扩张卷积(dilated convolutions),向卷积层引入了一个称为"扩张率(dilation rate)"的新参数,该参数定义了卷积核处理数据时各值的间距。
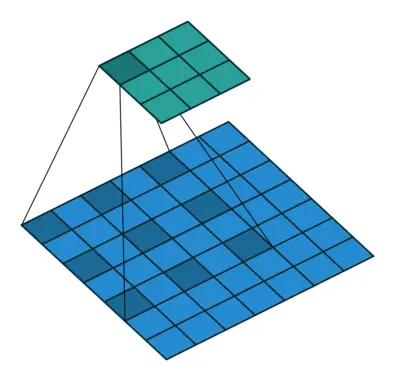
一个扩张率为2的3×3卷积核,感受野与5×5的卷积核相同,而且仅需要9个参数。你可以把它想象成一个5×5的卷积核,每隔一行或一列删除一行或一列。
在相同的计算条件下,空洞卷积提供了更大的感受野。空洞卷积经常用在实时图像分割中,当网络层需要较大的感受野,但计算资源有限而无法提高卷积核数量或大小时,可以考虑空洞卷积。
2.2.3 转置卷积(去卷积)
转置卷积(transposed Convolutions)又名反卷积(deconvolution)或是分数步长卷积(fractially straced convolutions_。
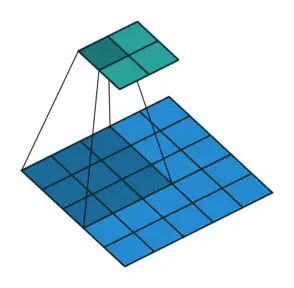
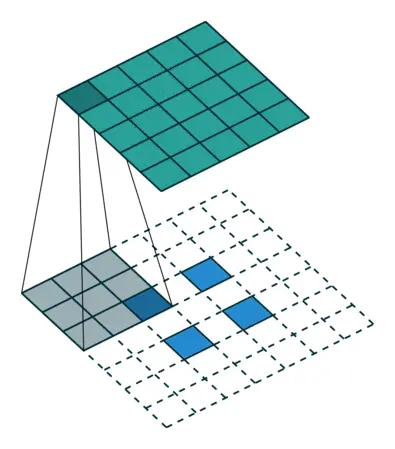
转置卷积的理解:
将一个普通卷积操作用一个矩阵表示,这个表示很简单,无非就是将卷积核重新排列到我们可以用普通的矩阵乘法进行矩阵卷积操作。如下图就是原始的卷积核:
我们对这个3×3的卷积核进行重新排列,得到了下面这个4×16的卷积矩阵:
这个便是卷积矩阵了,这个矩阵的每一行都定义了一个卷积操作。下图将会更加直观地告诉你这个重排列是怎么进行的,每一个卷积矩阵的行都是通过重新排列卷积核的元素,并且添加0补充(zero padding)进行的。
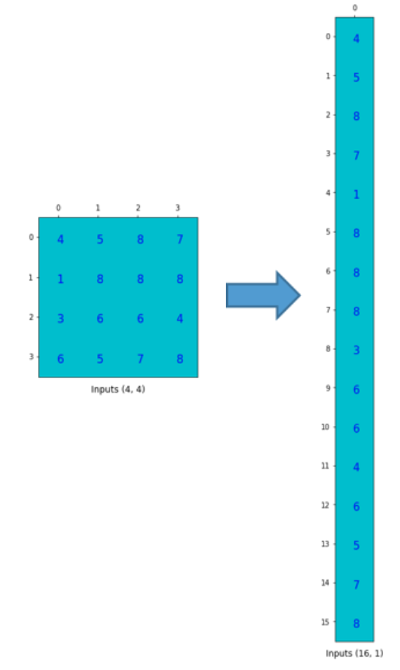
为了将卷积操作表示为卷积矩阵和输入矩阵的向量乘法,我们将输入矩阵4×4摊平(flatten)为一个列向量,形状为16×1,如下图所示。
我们可以将这个4×16的卷积矩阵和1×16的输入列向量进行矩阵乘法,这样我们就得到了输出列向量。
这个输出的4×1的矩阵可以重新塑性为一个2×2的矩阵,而这个矩阵正是和我们一开始通过传统的卷积操作得到的一模一样。
现在想要从4(2×2)到16(4×4),因此我们使用了一个16×4的矩阵,但是还有一件事情需要注意,想要维护一个1到9的映射关系。
假设我们转置这个卷积矩阵C(4×16)变为C^T(16\times 4)。我们可以对C^T和列向量(4×1)进行矩阵乘法,从而生成一个16×1的输出矩阵。这个转置矩阵正是将一个元素映射到了9个元素。
这个输出可以塑形为(4×4)的矩阵:
转置矩阵的算术解释可参阅:A guide to convolution arithmetic for deep learning
2.2 优化算法
https://zhuanlan.zhihu.com/p/81020717
https://zhuanlan.zhihu.com/p/43506482
https://zhuanlan.zhihu.com/p/81020717
https://zhuanlan.zhihu.com/p/41799394
- SGD
SGD是最基本的优化算法, 公式如下:
随机梯度优化算法有三个处理方式: 一次一个样本进行更新, 一次全部样本进行平均更新, 一次小批量进行平均更新的。
- Momentum
Momentum,也就是动量的意思。该算法将梯度下降的过程视为一个物理系统,下图是在百度图片中找的(侵删)
图片来自网络 如上图所示,在该物理系统中有一个小球(质点),它所处的水平方向的位置对应为 [公式] 的值,而垂直方向对应为损失。设其质量 [公式] ,在第 [公式] 时刻,在单位时间内,该质点受外力而造成的动量改变为:
[公式]
(1.1)到(1.2)是因为 [公式] ,所以约去了。另外受到的外力可以分为两个分量:重力沿斜面向下的力 [公式] 和粘性阻尼力 [公式]
[公式]
令
[公式]
代入(1.2)式中:
[公式]
然后对“位置”进行更新:
[公式]
所以这里 [公式] ,另外 [公式] 的方向与损失的梯度方向相反,并取系数为 [公式] ,得到:
[公式]
代入(1.4),得到速度的更新公式:
[公式]
进一步的,将(1.6)式展开,可以得到:
[公式]
可以看出来是一个变相的等比数列之和,且公比小于1,所以存在极限,当 [公式] 足够大时, [公式] 趋近于 [公式]
实现代码
import numpy as np
class Momentum(object): def init(self, alpha=0.9, lr=1e-3): self.alpha = alpha # 动量系数 self.lr = lr # 学习率 self.v = 0 # 初始速度为0
def update(self, g: np.ndarray): # g = J'(w) 为本轮训练参数的梯度
self.v = self.alpha * self.v - self.lr * g # 公式
return self.v # 返回的是参数的增量,下同
以上是基于指数衰减的实现方式,另外有的Momentum算法中会使用指数加权平均来实现,主要公式如下:
[公式]
不过该方式因为 [公式] ,刚开始时 [公式] 会比期望值要小,需要进行修正,下面的Adam等算法会使用该方式
Nesterov Momentum Nesterov Momentum是Momentum的改进版本,与Momentum唯一区别就是,Nesterov先用当前的速度 [公式] 更新一遍参数,得到一个临时参数 [公式] ,然后使用这个临时参数计算本轮训练的梯度。相当于是小球预判了自己下一时刻的位置,并提前使用该位置的梯度更新 :
[公式]
为了更加直观,还是上几个图吧,以下是Momentum算法 [公式] 的更新过程:
假设下一个位置的梯度如下:
那么Nesterov Momentum就提前使用这个梯度进行更新:
整体来看Nesterov的表现要好于Momentum,至于代码实现的话因为主要变化的是 [公式] ,所以可以之前使用Momentum的代码
AdaGrad AdaGrad全称为Adaptive Subgradient,其主要特点在于不断累加每次训练中梯度的平方,公式如下:
[公式]
其中 [公式] 是一个极小的正数,用来防止除0,而 [公式] , [公式] 是矩阵的哈达玛积运算符,另外,本文中矩阵的平方或者两矩阵相乘都是计算哈达玛积,而不是计算矩阵乘法
从公式中可以看出,随着算法不断迭代, [公式] 会越来越大,整体的学习率会越来越小。所以,一般来说AdaGrad算法一开始是激励收敛,到了后面就慢慢变成惩罚收敛,速度越来越慢
对于代码实现,首先将 [公式] 展开得到:
[公式]
通常 [公式] ,所以在第一次训练时(2.2)式为:
[公式]
因为每次训练 [公式] 的值是不确定的,所以要防止处0,但是可以令 [公式] ,这样就可以在(2.2)式中去掉 [公式] [公式]
将 [公式] 代入(2.3)式,可以得到:
[公式]
可知 [公式] 恒大于0,因此不必在计算 [公式] 中额外加入 [公式] ,代码如下:
class AdaGrad(object): def init(self, eps=1e-8, lr=1e-3): self.r = eps # r_0 = epsilon self.lr = lr
def update(self, g: np.ndarray):
r = r + np.square(g)
return -self.lr * g / np.sqrt(r)
RMSProp RMSProp是AdaGrad的改进算法,其公式和AdaGrad的区别只有 [公式] 的计算不同,先看公式
[公式]
可以看出,与AdaGrad不同,RMSProp只会累积近期的梯度信息,对于“遥远的历史”会以指数衰减的形式放弃
并且AdaGrad算法虽然在凸函数(Convex Functions)上表现较好,但是当目标函数非凸时,算法梯度下降的轨迹所经历的结构会复杂的多,早期梯度对当前训练没有太多意义,此时RMSProp往往表现更好
以下是将 [公式] 展开后的公式:
[公式]
与AdaGrad一样,令 [公式] ,从而去掉计算 [公式] 时的 [公式] ,实现代码:
class RMSProp(object): def init(self, lr=1e-3, beta=0.999, eps=1e-8): self.r = eps self.lr = lr self.beta = beta
def update(self, g: np.ndarray):
r = r * self.beta + (1-self.beta) * np.square(g)
return -self.lr * g / np.sqrt(r)
AdaDelta AdaDelta是与RMSProp相同时间对立发展出来的一个算法,在实现上可以看作是RMSProp的一个变种,先看公式:
[公式]
可以看到该算法不需要设置学习率 [公式] ,这是该算法的一大优势。除了同样以 [公式] 来累积梯度的信息之外,该算法还多了一个 [公式] 以指数衰减的形式来累积 [公式] 的信息
与前面相同,令:
[公式]
然后去掉(3.1)中的 [公式] ,得到:
[公式]
这样的话可以减少一些计算,代码如下:
class AdaDelta(object): def init(self, beta=0.999, eps=1e-8): self.r = eps self.s = eps self.beta = beta
def update(self, g: np.ndarray):
g_square = (1-self.beta) * np.square(g) # (1-beta)*g^2
r = r * self.beta + g_square
frac = s / r
res = -np.sqrt(frac) * g
s = s * self.beta + frac * g_squaretmp # 少一次乘法。。。
return res
关于以上几个算法的对比:
其中NAG是Nesterov Momentum
更多关于AdaDelta的信息,可以参考这篇文章:自适应学习率调整:AdaDelta
Adam Adam的名称来自Adaptive Momentum,可以看作是Momentum与RMSProp的一个结合体,该算法通过计算梯度的一阶矩估计和二阶矩估计而为不同的参数设计独立的自适应性学习率,公式如下:
[公式]
(4.1)和(4.2)在Momentum和RMSProp中已经介绍过了,而不直接使用 [公式] 计算 [公式] 却先经过(4.3)和(4.4)式是因为通常会设 [公式] ,所以此时梯度的一阶矩估计和二阶矩估是有偏的,需要进行修正
虽然没办法避免修正计算,但是还是可以省去一些计算过程,初始化时令:
[公式]
然后(4.5)式变为:
[公式]
因为 [公式] ,可知当 [公式] 足够大时修正将不起作用(也不需要修正了):
[公式]
代码如下:
class Adam(object): def init(self, lr=1e-3, alpha=0.9, beta=0.999, eps=1e-8): self.s = 0 self.r = eps self.lr = lr self.alpha = alpha self.beta = beta self.alpha_i = 1 self.beta_i = 1
def update(self, g: np.ndarray):
self.s = self.s * self.alpha + (1-self.alpha) * g
self.r = self.r * self.beta + (1-self.beta) * np.square(g)
self.alpha_i *= self.alpha
self.beta_i *= self.beta_i
lr = -self.lr * (1-self.beta_i)**0.5 / (1-self.alpha_i)
return lr * self.s / np.sqrt(self.r)
AdaMax 首先回顾RSMSProp中 [公式] 的展开式并且令 [公式] ,得到:
[公式]
可以看到这相当于是一个 [公式] 的 [公式] 范数,也就是说 [公式] 的各维度的增量是根据该维度上梯度的 [公式] 范数的累积量进行缩放的。如果用 [公式] 范数替代就得到了Adam的不同变种,不过其中 [公式] 范数对应的变种算法简单且稳定
对于 [公式] 范数,第 [公式] 轮训练时梯度的累积为:
[公式]
然后求无穷范数:
[公式]
由此再来递推 [公式] :
[公式]
需要注意,这个max比较的是梯度各个维度上的当前值和历史最大值,具体可以结合代码来看,最后其公式总结如下:
[公式]
另外,因为 [公式] 是累积的梯度各个分量的绝对值最大值,所以直接用做分母且不需要修正,代码如下:
class AdaMax(object): def init(self, lr=1e-3, alpha=0.9, beta=0.999): self.s = 0 self.r = 0 self.lr = lr self.alpha = alpha self.alpha_i = 1 self.beta = beta
def update(self, g: np.ndarray):
self.s = self.s * self.alpha + (1-self.alpha) * g
self.r = np.maximum(self.r*self.beta, np.abs(g))
self.alpha_i *= self.alpha
lr = -self.lr / (1-self.alpha_i)
return lr * self.s / self.r
Nadam Adam可以看作是Momentum与RMSProp的结合,既然Nesterov的表现较Momentum更优,那么自然也就可以把Nesterov Momentum与RMSProp组合到一起了,首先来看Nesterov的主要公式:
[公式]
为了令其更加接近Momentum,将(5.1)和(5.2)修改为:
[公式]
然后列出Adam中Momentum的部分:
[公式]
将(5.5)和(5.6)式代入到(5.7)式中:
[公式]
将上式中标红部分进行近似:
[公式]
代入原式,得到:
[公式]
接着,按照(5.4)式的套路,将 [公式] 替换成 [公式] ,得到:
[公式]
整理一下公式:
[公式]
同样令 [公式] ,消去(5.8)式种的 [公式] :
[公式]
代码
class Nadam(object): def init(self, lr=1e-3, alpha=0.9, beta=0.999, eps=1e-8): self.s = 0 self.r = eps self.lr = lr self.alpha = alpha self.beta = beta self.alpha_i = 1 self.beta_i = 1
def update(self, g: np.ndarray):
self.s = self.s * self.alpha + (1-self.alpha) * g
self.r = self.r * self.beta + (1-self.beta) * np.square(g)
self.alpha_i *= self.alpha
self.beta_i *= self.beta_i
lr = -self.lr * (1-self.beta_i)**0.5 / (1-self.alpha_i)
return lr * (self.s * self.alpha + (1-self.alpha) * g) / np.sqrt(self.r)
NadaMax 按照同样的思路,可以将Nesterov与AdaMax结合变成NadaMax,回顾以下(5.8)式:
[公式]
然后是AdaMax的二阶矩估计部分:
[公式]
用(6.2)式替换掉(6.1)式中标红部分,得到:
[公式]
最后,整理公式:
[公式]
代码实现:
class NadaMax(object): def init(self, lr=1e-3, alpha=0.9, beta=0.999): self.s = 0 self.r = 0 self.lr = lr self.alpha = alpha self.alpha_i = 1 self.beta = beta
def update(self, g: np.ndarray):
self.s = self.s * self.alpha + (1-self.alpha) * g
self.r = np.maximum(self.r*self.beta, np.abs(g))
self.alpha_i *= self.alpha
lr = -self.lr / (1-self.alpha_i)
return lr * (self.s * self.alpha + (1-self.alpha) * g) / self.r
初始化
https://www.zhihu.com/question/291032522/answer/605843215
https://www.zhihu.com/question/312556066/answer/600228264
https://www.zhihu.com/question/291032522/answer/605843215
https://cloud.tencent.com/developer/article/1352583
https://zhuanlan.zhihu.com/p/44106492
三、经典网络
https://zhuanlan.zhihu.com/p/66215918
https://zhuanlan.zhihu.com/p/61185030
https://zhuanlan.zhihu.com/p/60479586
https://zhuanlan.zhihu.com/p/60187262
https://zhuanlan.zhihu.com/p/64693337
https://blog.csdn.net/SIGAI_CSDN/article/details/85098096
https://zhuanlan.zhihu.com/p/57399396
3.1 Lenet网络
参考论文: Gradient-Based Learning Applied to Document Recognition
卷积神经网络的开山之作,麻雀虽小五脏俱全,卷积层、池化层、全链接层一直沿用至今。

网络特点:
相比MLP,LeNet使用了相对更少的参数,获得了更好的结果。
设计了maxpool来提取特征
# Contains a variant of the LeNet model definition.
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v1 as tf
import tf_slim as slim
def lenet(images, num_classes=10, is_training=False, dropout_keep_prob=0.5,
prediction_fn=slim.softmax, scope='LeNet'):
"""
Creates a variant of the LeNet model.
Note that since the output is a set of 'logits', the values fall in the
interval of (-infinity, infinity). Consequently, to convert the outputs to a
probability distribution over the characters, one will need to convert them
using the softmax function:
logits = lenet.lenet(images, is_training=False)
probabilities = tf.nn.softmax(logits)
predictions = tf.argmax(logits, 1)
Args:
images: A batch of `Tensors` of size [batch_size, height, width, channels].
num_classes: the number of classes in the dataset. If 0 or None, the logits
layer is omitted and the input features to the logits layer are returned instead.
is_training: specifies whether or not we're currently training the model.
This variable will determine the behaviour of the dropout layer.
dropout_keep_prob: the percentage of activation values that are retained.
prediction_fn: a function to get predictions out of logits.
scope: Optional variable_scope.
Returns:
net: a 2D Tensor with the logits (pre-softmax activations) if num_classes
is a non-zero integer, or the inon-dropped-out nput to the logits layer
if num_classes is 0 or None.
end_points: a dictionary from components of the network to the corresponding
activation.
"""
end_points = {}
with tf.variable_scope(scope, 'LeNet', [images]):
net = end_points['conv1'] = slim.conv2d(images, 32, [5, 5], scope='conv1')
net = end_points['pool1'] = slim.max_pool2d(net, [2, 2], 2, scope='pool1')
net = end_points['conv2'] = slim.conv2d(net, 64, [5, 5], scope='conv2')
net = end_points['pool2'] = slim.max_pool2d(net, [2, 2], 2, scope='pool2')
net = slim.flatten(net)
end_points['Flatten'] = net
net = end_points['fc3'] = slim.fully_connected(net, 1024, scope='fc3')
if not num_classes:
return net, end_points
net = end_points['dropout3'] = slim.dropout(net, dropout_keep_prob, is_training=is_training, scope='dropout3')
logits = end_points['Logits'] = slim.fully_connected(net, num_classes, activation_fn=None, scope='fc4')
end_points['Predictions'] = prediction_fn(logits, scope='Predictions')
return logits, end_points
lenet.default_image_size = 28
def lenet_arg_scope(weight_decay=0.0):
"""
Defines the default lenet argument scope.
Args:
weight_decay: The weight decay to use for regularizing the model.
Returns:
An `arg_scope` to use for the inception v3 model.
"""
with slim.arg_scope([slim.conv2d, slim.fully_connected],
weights_regularizer=slim.l2_regularizer(weight_decay),
weights_initializer=tf.truncated_normal_initializer(stddev=0.1),
activation_fn=tf.nn.relu) as sc:
return sc
3.2 Alexnet网络
AlexNet是Hinton和他的学生Alex在2012设计的网络,并获得了当年的ImageNet竞赛冠军。参考论文: ImageNet Classification with Deep Convolutional Neural Networks
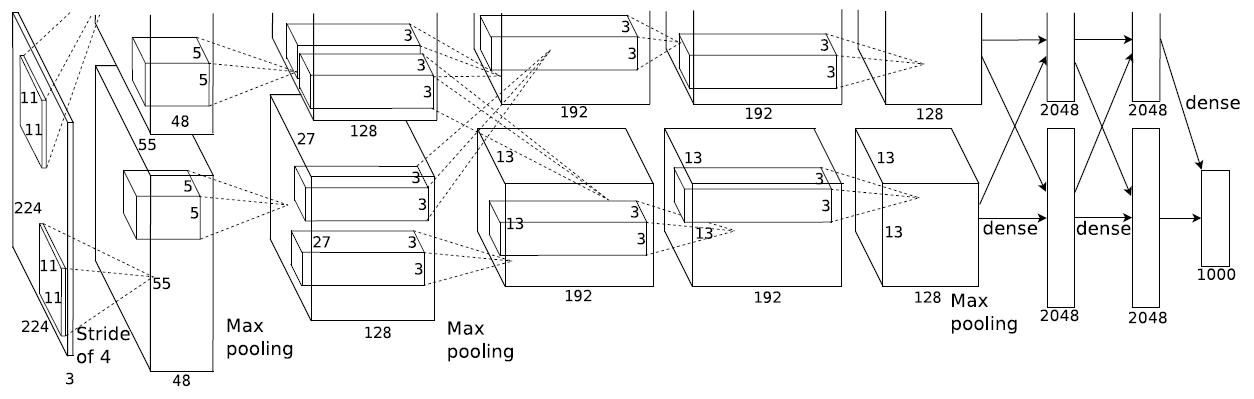
AlexNet主要使用到的新技术点如下:
- 成功使用ReLU作为CNN的激活函数,并验证其效果在较深的网络超过了Sigmoid,成功解决了Sigmoid在网络较深时的梯度弥散问题。虽然ReLU激活函数在很久之前就被提出了,但是直到AlexNet的出现才将其发扬光大。
- 训练时使用Dropout随机忽略一部分神经元,以避免模型过拟合。Dropout虽有单独的论文论述,但是AlexNet将其实用化,通过实践证实了它的效果。在AlexNet中主要是最后几个全连接层使用了Dropout。
- 在CNN中使用重叠的最大池化。此前CNN中普遍使用平均池化,AlexNet全部使用最大池化,避免平均池化的模糊化效果。并且AlexNet中提出让步长比池化核的尺寸小,这样池化层的输出之间会有重叠和覆盖,提升了特征的丰富性。
- 提出了LRN层,对局部神经元的活动创建竞争机制,使得其中响应比较大的值变得相对更大,并抑制其他反馈较小的神经元,增强了模型的泛化能力。
- 相比LeNet,AlexNet设计了更深层的网络。
- 通过裁剪,旋转等方式增强了训练数据。
- 受于当时的算力限制,Alexnet创新地将图像分为上下两块分别训练,然后在全连接层合并在一起(AlexNet网络图,可以看到有上下两部分)。
# -*- coding:utf8 -*-
"""
Contains a model definition for AlexNet.
This work was first described in:
ImageNet Classification with Deep Convolutional Neural Networks
Alex Krizhevsky, Ilya Sutskever and Geoffrey E. Hinton
and later refined in:
One weird trick for parallelizing convolutional neural networks
Alex Krizhevsky, 2014
Here we provide the implementation proposed in "One weird trick" and not
"ImageNet Classification", as per the paper, the LRN layers have been removed.
Usage:
with slim.arg_scope(alexnet.alexnet_v2_arg_scope()):
outputs, end_points = alexnet.alexnet_v2(inputs)
"""
import tensorflow.compat.v1 as tf
import tf_slim as slim
# pylint: disable=g-long-lambda
trunc_normal = lambda stddev: tf.truncated_normal_initializer(0.0, stddev)
def alexnet_v2_arg_scope(weight_decay=0.0005):
with slim.arg_scope([slim.conv2d, slim.fully_connected],
activation_fn=tf.nn.relu,
biases_initializer=tf.constant_initializer(0.1),
weights_regularizer=slim.l2_regularizer(weight_decay)):
with slim.arg_scope([slim.conv2d], padding='SAME'):
with slim.arg_scope([slim.max_pool2d], padding='VALID') as arg_sc:
return arg_sc
def alexnet_v2(inputs,num_classes=1000, is_training=True,dropout_keep_prob=0.5,
spatial_squeeze=True,scope='alexnet_v2',global_pool=False):
"""
AlexNet version 2.
Described in: http://arxiv.org/pdf/1404.5997v2.pdf
Parameters from: github.com/akrizhevsky/cuda-convnet2/blob/master/layers/layers-imagenet-1gpu.cfg
Note: All the fully_connected layers have been transformed to conv2d layers.
To use in classification mode, resize input to 224x224 or set
global_pool=True. To use in fully convolutional mode, set
spatial_squeeze to false.
The LRN layers have been removed and change the initializers from
random_normal_initializer to xavier_initializer.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
num_classes: the number of predicted classes. If 0 or None, the logits layer
is omitted and the input features to the logits layer are returned instead.
is_training: whether or not the model is being trained.
dropout_keep_prob: the probability that activations are kept in the dropout
layers during training.
spatial_squeeze: whether or not should squeeze the spatial dimensions of the
logits. Useful to remove unnecessary dimensions for classification.
scope: Optional scope for the variables.
global_pool: Optional boolean flag. If True, the input to the classification
layer is avgpooled to size 1x1, for any input size. (This is not part
of the original AlexNet.)
Returns:
net: the output of the logits layer (if num_classes is a non-zero integer),
or the non-dropped-out input to the logits layer (if num_classes is 0
or None).
end_points: a dict of tensors with intermediate activations.
"""
with tf.variable_scope(scope, 'alexnet_v2', [inputs]) as sc:
end_points_collection = sc.original_name_scope + '_end_points'
# Collect outputs for conv2d, fully_connected and max_pool2d.
with slim.arg_scope([slim.conv2d, slim.fully_connected, slim.max_pool2d],outputs_collections=[end_points_collection]):
net = slim.conv2d(inputs, 64, [11, 11], 4, padding='VALID',scope='conv1')
net = slim.max_pool2d(net, [3, 3], 2, scope='pool1')
net = slim.conv2d(net, 192, [5, 5], scope='conv2')
net = slim.max_pool2d(net, [3, 3], 2, scope='pool2')
net = slim.conv2d(net, 384, [3, 3], scope='conv3')
net = slim.conv2d(net, 384, [3, 3], scope='conv4')
net = slim.conv2d(net, 256, [3, 3], scope='conv5')
net = slim.max_pool2d(net, [3, 3], 2, scope='pool5')
# Use conv2d instead of fully_connected layers.
with slim.arg_scope([slim.conv2d],weights_initializer=trunc_normal(0.005),biases_initializer=tf.constant_initializer(0.1)):
net = slim.conv2d(net, 4096, [5, 5], padding='VALID',scope='fc6')
net = slim.dropout(net, dropout_keep_prob, is_training=is_training,scope='dropout6')
net = slim.conv2d(net, 4096, [1, 1], scope='fc7')
# Convert end_points_collection into a end_point dict.
end_points = slim.utils.convert_collection_to_dict(end_points_collection)
if global_pool:
net = tf.reduce_mean(input_tensor=net, axis=[1, 2], keepdims=True, name='global_pool')
end_points['global_pool'] = net
if num_classes:
net = slim.dropout(net, dropout_keep_prob, is_training=is_training,scope='dropout7')
net = slim.conv2d(net,num_classes, [1, 1],activation_fn=None,normalizer_fn=None,biases_initializer=tf.zeros_initializer(),scope='fc8')
if spatial_squeeze:
net = tf.squeeze(net, [1, 2], name='fc8/squeezed')
end_points[sc.name + '/fc8'] = net
return net, end_points
alexnet_v2.default_image_size = 224
3.3 VGG网络
参考论文: Very Deep Convolutional Networks for Large-Scale Image Recognition
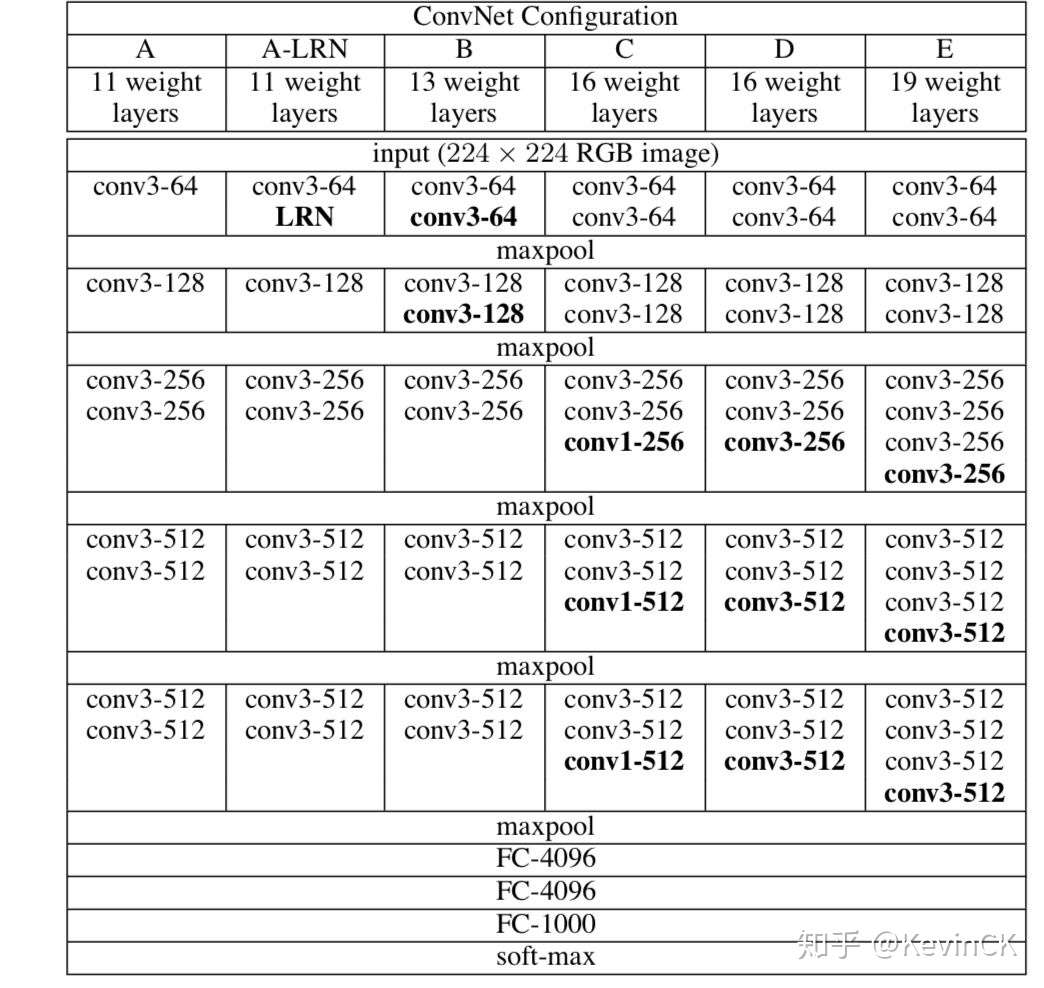
VGG优点:
VGGNet的结构非常简洁,整个网络都使用了同样大小的卷积核尺寸(3x3)和最大池化尺寸(2x2)。
几个小滤波器(3x3)卷积层的组合比一个大滤波器(5x5或7x7)卷积层好
验证了通过不断加深网络结构可以提升性能。
VGG缺点
VGG耗费更多计算资源,并且使用了更多的参数(这里不是3x3卷积的锅)m导致更多的内存占用(140M)。其中绝大多数的参数都是来自于第一个全连接层。VGG可是有3个全连接层啊!
# -*- coding:utf8 -*-
"""
Contains model definitions for versions of the Oxford VGG network.
These model definitions were introduced in the following technical report:
Very Deep Convolutional Networks For Large-Scale Image Recognition
Karen Simonyan and Andrew Zisserman
PDF: http://arxiv.org/pdf/1409.1556.pdf
ILSVRC 2014 Slides: http://www.robots.ox.ac.uk/~karen/pdf/ILSVRC_2014.pdf
More information can be obtained from the VGG website:
www.robots.ox.ac.uk/~vgg/research/very_deep/
Usage:
with slim.arg_scope(vgg.vgg_arg_scope()):
outputs, end_points = vgg.vgg_a(inputs)
with slim.arg_scope(vgg.vgg_arg_scope()):
outputs, end_points = vgg.vgg_16(inputs)
"""
import tensorflow.compat.v1 as tf
import tf_slim as slim
def vgg_arg_scope(weight_decay=0.0005):
"""
Defines the VGG arg scope.
Args:
weight_decay: The l2 regularization coefficient.
Returns:
An arg_scope.
"""
with slim.arg_scope([slim.conv2d, slim.fully_connected], activation_fn=tf.nn.relu,
weights_regularizer=slim.l2_regularizer(weight_decay),
biases_initializer=tf.zeros_initializer()):
with slim.arg_scope([slim.conv2d], padding='SAME') as arg_sc:
return arg_sc
def vgg_a(inputs,num_classes=1000, is_training=True,dropout_keep_prob=0.5,spatial_squeeze=True,reuse=None,
scope='vgg_a',fc_conv_padding='VALID',global_pool=False):
"""
Oxford Net VGG 11-Layers version A Example.
Note: All the fully_connected layers have been transformed to conv2d layers.
To use in classification mode, resize input to 224x224.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
num_classes: number of predicted classes. If 0 or None, the logits layer is
omitted and the input features to the logits layer are returned instead.
is_training: whether or not the model is being trained.
dropout_keep_prob: the probability that activations are kept in the dropout
layers during training.
spatial_squeeze: whether or not should squeeze the spatial dimensions of the
outputs. Useful to remove unnecessary dimensions for classification.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
scope: Optional scope for the variables.
fc_conv_padding: the type of padding to use for the fully connected layer
that is implemented as a convolutional layer. Use 'SAME' padding if you
are applying the network in a fully convolutional manner and want to
get a prediction map downsampled by a factor of 32 as an output.
Otherwise, the output prediction map will be (input / 32) - 6 in case of
'VALID' padding.
global_pool: Optional boolean flag. If True, the input to the classification
layer is avgpooled to size 1x1, for any input size. (This is not part
of the original VGG architecture.)
Returns:
net: the output of the logits layer (if num_classes is a non-zero integer),
or the input to the logits layer (if num_classes is 0 or None).
end_points: a dict of tensors with intermediate activations.
"""
with tf.variable_scope(scope, 'vgg_a', [inputs], reuse=reuse) as sc:
end_points_collection = sc.original_name_scope + '_end_points'
# Collect outputs for conv2d, fully_connected and max_pool2d.
with slim.arg_scope([slim.conv2d, slim.max_pool2d],outputs_collections=end_points_collection):
net = slim.repeat(inputs, 1, slim.conv2d, 64, [3, 3], scope='conv1')
net = slim.max_pool2d(net, [2, 2], scope='pool1')
net = slim.repeat(net, 1, slim.conv2d, 128, [3, 3], scope='conv2')
net = slim.max_pool2d(net, [2, 2], scope='pool2')
net = slim.repeat(net, 2, slim.conv2d, 256, [3, 3], scope='conv3')
net = slim.max_pool2d(net, [2, 2], scope='pool3')
net = slim.repeat(net, 2, slim.conv2d, 512, [3, 3], scope='conv4')
net = slim.max_pool2d(net, [2, 2], scope='pool4')
net = slim.repeat(net, 2, slim.conv2d, 512, [3, 3], scope='conv5')
net = slim.max_pool2d(net, [2, 2], scope='pool5')
# Use conv2d instead of fully_connected layers.
net = slim.conv2d(net, 4096, [7, 7], padding=fc_conv_padding, scope='fc6')
net = slim.dropout(net, dropout_keep_prob, is_training=is_training, scope='dropout6')
net = slim.conv2d(net, 4096, [1, 1], scope='fc7')
# Convert end_points_collection into a end_point dict.
end_points = slim.utils.convert_collection_to_dict(end_points_collection)
if global_pool:
net = tf.reduce_mean(input_tensor=net, axis=[1, 2], keepdims=True, name='global_pool')
end_points['global_pool'] = net
if num_classes:
net = slim.dropout(net, dropout_keep_prob, is_training=is_training, scope='dropout7')
net = slim.conv2d(net, num_classes, [1, 1], activation_fn=None,normalizer_fn=None, scope='fc8')
if spatial_squeeze:
net = tf.squeeze(net, [1, 2], name='fc8/squeezed')
end_points[sc.name + '/fc8'] = net
return net, end_points
vgg_a.default_image_size = 224
def vgg_16(inputs,num_classes=1000,is_training=True,dropout_keep_prob=0.5,spatial_squeeze=True,
reuse=None,scope='vgg_16',fc_conv_padding='VALID',global_pool=False):
"""
Oxford Net VGG 16-Layers version D Example.
Note: All the fully_connected layers have been transformed to conv2d layers.
To use in classification mode, resize input to 224x224.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
num_classes: number of predicted classes. If 0 or None, the logits layer is
omitted and the input features to the logits layer are returned instead.
is_training: whether or not the model is being trained.
dropout_keep_prob: the probability that activations are kept in the dropout
layers during training.
spatial_squeeze: whether or not should squeeze the spatial dimensions of the
outputs. Useful to remove unnecessary dimensions for classification.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
scope: Optional scope for the variables.
fc_conv_padding: the type of padding to use for the fully connected layer
that is implemented as a convolutional layer. Use 'SAME' padding if you
are applying the network in a fully convolutional manner and want to
get a prediction map downsampled by a factor of 32 as an output.
Otherwise, the output prediction map will be (input / 32) - 6 in case of
'VALID' padding.
global_pool: Optional boolean flag. If True, the input to the classification
layer is avgpooled to size 1x1, for any input size. (This is not part
of the original VGG architecture.)
Returns:
net: the output of the logits layer (if num_classes is a non-zero integer),
or the input to the logits layer (if num_classes is 0 or None).
end_points: a dict of tensors with intermediate activations.
"""
with tf.variable_scope(scope, 'vgg_16', [inputs], reuse=reuse) as sc:
end_points_collection = sc.original_name_scope + '_end_points'
# Collect outputs for conv2d, fully_connected and max_pool2d.
with slim.arg_scope([slim.conv2d, slim.fully_connected, slim.max_pool2d], outputs_collections=end_points_collection):
net = slim.repeat(inputs, 2, slim.conv2d, 64, [3, 3], scope='conv1')
net = slim.max_pool2d(net, [2, 2], scope='pool1')
net = slim.repeat(net, 2, slim.conv2d, 128, [3, 3], scope='conv2')
net = slim.max_pool2d(net, [2, 2], scope='pool2')
net = slim.repeat(net, 3, slim.conv2d, 256, [3, 3], scope='conv3')
net = slim.max_pool2d(net, [2, 2], scope='pool3')
net = slim.repeat(net, 3, slim.conv2d, 512, [3, 3], scope='conv4')
net = slim.max_pool2d(net, [2, 2], scope='pool4')
net = slim.repeat(net, 3, slim.conv2d, 512, [3, 3], scope='conv5')
net = slim.max_pool2d(net, [2, 2], scope='pool5')
# Use conv2d instead of fully_connected layers.
net = slim.conv2d(net, 4096, [7, 7], padding=fc_conv_padding, scope='fc6')
net = slim.dropout(net, dropout_keep_prob, is_training=is_training, scope='dropout6')
net = slim.conv2d(net, 4096, [1, 1], scope='fc7')
# Convert end_points_collection into a end_point dict.
end_points = slim.utils.convert_collection_to_dict(end_points_collection)
if global_pool:
net = tf.reduce_mean(input_tensor=net, axis=[1, 2], keepdims=True, name='global_pool')
end_points['global_pool'] = net
if num_classes:
net = slim.dropout(net, dropout_keep_prob, is_training=is_training, scope='dropout7')
net = slim.conv2d(net, num_classes, [1, 1],activation_fn=None,normalizer_fn=None,scope='fc8')
if spatial_squeeze:
net = tf.squeeze(net, [1, 2], name='fc8/squeezed')
end_points[sc.name + '/fc8'] = net
return net, end_points
vgg_16.default_image_size = 224
def vgg_19(inputs,num_classes=1000,is_training=True,dropout_keep_prob=0.5,spatial_squeeze=True,
reuse=None,scope='vgg_19',fc_conv_padding='VALID',global_pool=False):
"""
Oxford Net VGG 19-Layers version E Example.
Note: All the fully_connected layers have been transformed to conv2d layers.
To use in classification mode, resize input to 224x224.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
num_classes: number of predicted classes. If 0 or None, the logits layer is
omitted and the input features to the logits layer are returned instead.
is_training: whether or not the model is being trained.
dropout_keep_prob: the probability that activations are kept in the dropout
layers during training.
spatial_squeeze: whether or not should squeeze the spatial dimensions of the
outputs. Useful to remove unnecessary dimensions for classification.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
scope: Optional scope for the variables.
fc_conv_padding: the type of padding to use for the fully connected layer
that is implemented as a convolutional layer. Use 'SAME' padding if you
are applying the network in a fully convolutional manner and want to
get a prediction map downsampled by a factor of 32 as an output.
Otherwise, the output prediction map will be (input / 32) - 6 in case of
'VALID' padding.
global_pool: Optional boolean flag. If True, the input to the classification
layer is avgpooled to size 1x1, for any input size. (This is not part
of the original VGG architecture.)
Returns:
net: the output of the logits layer (if num_classes is a non-zero integer),
or the non-dropped-out input to the logits layer (if num_classes is 0 or
None).
end_points: a dict of tensors with intermediate activations.
"""
with tf.variable_scope(scope, 'vgg_19', [inputs], reuse=reuse) as sc:
end_points_collection = sc.original_name_scope + '_end_points'
# Collect outputs for conv2d, fully_connected and max_pool2d.
with slim.arg_scope([slim.conv2d, slim.fully_connected, slim.max_pool2d],outputs_collections=end_points_collection):
net = slim.repeat(inputs, 2, slim.conv2d, 64, [3, 3], scope='conv1')
net = slim.max_pool2d(net, [2, 2], scope='pool1')
net = slim.repeat(net, 2, slim.conv2d, 128, [3, 3], scope='conv2')
net = slim.max_pool2d(net, [2, 2], scope='pool2')
net = slim.repeat(net, 4, slim.conv2d, 256, [3, 3], scope='conv3')
net = slim.max_pool2d(net, [2, 2], scope='pool3')
net = slim.repeat(net, 4, slim.conv2d, 512, [3, 3], scope='conv4')
net = slim.max_pool2d(net, [2, 2], scope='pool4')
net = slim.repeat(net, 4, slim.conv2d, 512, [3, 3], scope='conv5')
net = slim.max_pool2d(net, [2, 2], scope='pool5')
# Use conv2d instead of fully_connected layers.
net = slim.conv2d(net, 4096, [7, 7], padding=fc_conv_padding, scope='fc6')
net = slim.dropout(net, dropout_keep_prob, is_training=is_training,scope='dropout6')
net = slim.conv2d(net, 4096, [1, 1], scope='fc7')
# Convert end_points_collection into a end_point dict.
end_points = slim.utils.convert_collection_to_dict(end_points_collection)
if global_pool:
net = tf.reduce_mean(
input_tensor=net, axis=[1, 2], keepdims=True, name='global_pool')
end_points['global_pool'] = net
if num_classes:
net = slim.dropout(net, dropout_keep_prob, is_training=is_training,scope='dropout7')
net = slim.conv2d(net, num_classes, [1, 1],activation_fn=None,normalizer_fn=None,scope='fc8')
if spatial_squeeze:
net = tf.squeeze(net, [1, 2], name='fc8/squeezed')
end_points[sc.name + '/fc8'] = net
return net, end_points
vgg_19.default_image_size = 224
# Alias
vgg_d = vgg_16
vgg_e = vgg_19
3.4 GoogleNet
3.4.1 InceptionV1
参考论文: Going deeper with convolutions
inception模块:
网络结构:
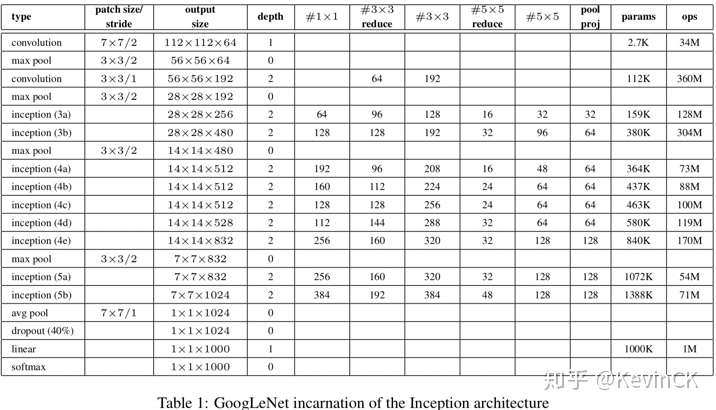
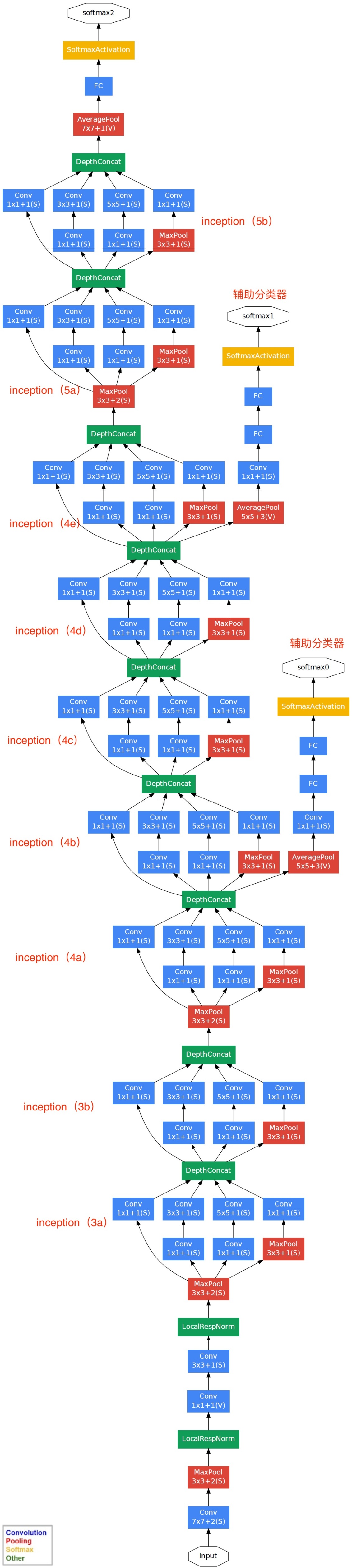
网络的特点:
- 引入Inception概念,人为构建稀疏连接,引入多尺度感受野和多尺度融合
- 采用Network in Network中用Average pool来代替全连接层的思想,大幅度减少参数数目和计算量,一定程度上引入了正则化,同时使得网络输入的尺寸可变。实际在最后一层还是添加了一个全连接层,是为了做finetune。
- 另外增加了两个辅助的softmax分支,作用有两点,一是为了避免梯度消失,用于向前传导梯度。反向传播时如果有一层求导为0,链式求导结果则为0。二是将中间某一层输出用作分类,起到模型融合作用。最后的loss=loss_2 + 0.3 * loss_1 + 0.3 * loss_0。实际测试时,这两个辅助softmax分支会被去掉。
- 在NIN模型中与1*1卷积层等效的MLPConv既能跨通道组织信息,提高网络的表达能力,同时可以对输出有效进行降维。
# -*- coding:utf -*-
# FILE inception_utils.py
"""
Contains common code shared by all inception models.
Usage of arg scope:
with slim.arg_scope(inception_arg_scope()):
logits, end_points = inception.inception_v3(images, num_classes, is_training=is_training)
"""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v1 as tf
import tf_slim as slim
def inception_arg_scope(weight_decay=0.00004, use_batch_norm=True, batch_norm_decay=0.9997, batch_norm_epsilon=0.001,
activation_fn=tf.nn.relu, batch_norm_updates_collections=tf.GraphKeys.UPDATE_OPS, batch_norm_scale=False):
"""
Defines the default arg scope for inception models.
Args:
weight_decay: The weight decay to use for regularizing the model.
use_batch_norm: "If `True`, batch_norm is applied after each convolution.
batch_norm_decay: Decay for batch norm moving average.
batch_norm_epsilon: Small float added to variance to avoid dividing by zero
in batch norm.
activation_fn: Activation function for conv2d.
batch_norm_updates_collections: Collection for the update ops for
batch norm.
batch_norm_scale: If True, uses an explicit `gamma` multiplier to scale the
activations in the batch normalization layer.
Returns:
An `arg_scope` to use for the inception models.
"""
batch_norm_params = {
# Decay for the moving averages.
'decay': batch_norm_decay,
# epsilon to prevent 0s in variance.
'epsilon': batch_norm_epsilon,
# collection containing update_ops.
'updates_collections': batch_norm_updates_collections,
# use fused batch norm if possible.
'fused': None,
'scale': batch_norm_scale,
}
if use_batch_norm:
normalizer_fn = slim.batch_norm
normalizer_params = batch_norm_params
else:
normalizer_fn = None
normalizer_params = {}
# Set weight_decay for weights in Conv and FC layers.
with slim.arg_scope([slim.conv2d, slim.fully_connected], weights_regularizer=slim.l2_regularizer(weight_decay)):
with slim.arg_scope([slim.conv2d], weights_initializer=slim.variance_scaling_initializer(),
activation_fn=activation_fn,normalizer_fn=normalizer_fn,normalizer_params=normalizer_params) as sc:
return sc
# -*- coding:utf8 -*-
# FIlE inception_v1.py
# Contains the definition for inception v1 classification network.
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v1 as tf
import tf_slim as slim
from nets import inception_utils
# pylint: disable=g-long-lambda
trunc_normal = lambda stddev: tf.truncated_normal_initializer(0.0, stddev)
def inception_v1_base(inputs, final_endpoint='Mixed_5c', include_root_block=True, scope='InceptionV1'):
"""
Defines the Inception V1 base architecture.
This architecture is defined in:
Going deeper with convolutions
Christian Szegedy, Wei Liu, Yangqing Jia, Pierre Sermanet, Scott Reed,
Dragomir Anguelov, Dumitru Erhan, Vincent Vanhoucke, Andrew Rabinovich.
http://arxiv.org/pdf/1409.4842v1.pdf.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
final_endpoint: specifies the endpoint to construct the network up to. It
can be one of ['Conv2d_1a_7x7', 'MaxPool_2a_3x3', 'Conv2d_2b_1x1',
'Conv2d_2c_3x3', 'MaxPool_3a_3x3', 'Mixed_3b', 'Mixed_3c',
'MaxPool_4a_3x3', 'Mixed_4b', 'Mixed_4c', 'Mixed_4d', 'Mixed_4e',
'Mixed_4f', 'MaxPool_5a_2x2', 'Mixed_5b', 'Mixed_5c']. If
include_root_block is False, ['Conv2d_1a_7x7', 'MaxPool_2a_3x3',
'Conv2d_2b_1x1', 'Conv2d_2c_3x3', 'MaxPool_3a_3x3'] will not be available.
include_root_block: If True, include the convolution and max-pooling layers
before the inception modules. If False, excludes those layers.
scope: Optional variable_scope.
Returns:
A dictionary from components of the network to the corresponding activation.
Raises:
ValueError: if final_endpoint is not set to one of the predefined values.
"""
end_points = {}
with tf.variable_scope(scope, 'InceptionV1', [inputs]):
with slim.arg_scope([slim.conv2d, slim.fully_connected], weights_initializer=trunc_normal(0.01)):
with slim.arg_scope([slim.conv2d, slim.max_pool2d],stride=1, padding='SAME'):
net = inputs
if include_root_block:
end_point = 'Conv2d_1a_7x7'
net = slim.conv2d(inputs, 64, [7, 7], stride=2, scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point:
return net, end_points
end_point = 'MaxPool_2a_3x3'
net = slim.max_pool2d(net, [3, 3], stride=2, scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point:
return net, end_points
end_point = 'Conv2d_2b_1x1'
net = slim.conv2d(net, 64, [1, 1], scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point:
return net, end_points
end_point = 'Conv2d_2c_3x3'
net = slim.conv2d(net, 192, [3, 3], scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point:
return net, end_points
end_point = 'MaxPool_3a_3x3'
net = slim.max_pool2d(net, [3, 3], stride=2, scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point:
return net, end_points
end_point = 'Mixed_3b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 64, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 96, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 128, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 16, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 32, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 32, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_3c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 128, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 128, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 192, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 32, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 96, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 64, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'MaxPool_4a_3x3'
net = slim.max_pool2d(net, [3, 3], stride=2, scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_4b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 192, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 96, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 208, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 16, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 48, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 64, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_4c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 160, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 112, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 224, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 24, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 64, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 64, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_4d'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 128, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 128, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 256, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 24, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 64, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 64, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_4e'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 112, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 144, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 288, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 32, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 64, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 64, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_4f'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 256, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 160, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 320, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 32, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 128, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 128, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'MaxPool_5a_2x2'
net = slim.max_pool2d(net, [2, 2], stride=2, scope=end_point)
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_5b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 256, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 160, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 320, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 32, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 128, [3, 3], scope='Conv2d_0a_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 128, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
end_point = 'Mixed_5c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 384, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 192, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 384, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, 48, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 128, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 128, [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if final_endpoint == end_point: return net, end_points
raise ValueError('Unknown final endpoint %s' % final_endpoint)
def inception_v1(inputs, num_classes=1000, is_training=True, dropout_keep_prob=0.8, prediction_fn=slim.softmax,
spatial_squeeze=True, reuse=None, scope='InceptionV1', global_pool=False):
"""
Defines the Inception V1 architecture.
This architecture is defined in:
Going deeper with convolutions
Christian Szegedy, Wei Liu, Yangqing Jia, Pierre Sermanet, Scott Reed,
Dragomir Anguelov, Dumitru Erhan, Vincent Vanhoucke, Andrew Rabinovich.
http://arxiv.org/pdf/1409.4842v1.pdf.
The default image size used to train this network is 224x224.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
num_classes: number of predicted classes. If 0 or None, the logits layer
is omitted and the input features to the logits layer (before dropout)
are returned instead.
is_training: whether is training or not.
dropout_keep_prob: the percentage of activation values that are retained.
prediction_fn: a function to get predictions out of logits.
spatial_squeeze: if True, logits is of shape [B, C], if false logits is of
shape [B, 1, 1, C], where B is batch_size and C is number of classes.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
scope: Optional variable_scope.
global_pool: Optional boolean flag to control the avgpooling before the
logits layer. If false or unset, pooling is done with a fixed window
that reduces default-sized inputs to 1x1, while larger inputs lead to
larger outputs. If true, any input size is pooled down to 1x1.
Returns:
net: a Tensor with the logits (pre-softmax activations) if num_classes
is a non-zero integer, or the non-dropped-out input to the logits layer
if num_classes is 0 or None.
end_points: a dictionary from components of the network to the corresponding
activation.
"""
# Final pooling and prediction
with tf.variable_scope(scope, 'InceptionV1', [inputs], reuse=reuse) as scope:
with slim.arg_scope([slim.batch_norm, slim.dropout],is_training=is_training):
net, end_points = inception_v1_base(inputs, scope=scope)
with tf.variable_scope('Logits'):
if global_pool:
# Global average pooling.
net = tf.reduce_mean(input_tensor=net, axis=[1, 2], keepdims=True, name='global_pool')
end_points['global_pool'] = net
else:
# Pooling with a fixed kernel size.
net = slim.avg_pool2d(net, [7, 7], stride=1, scope='AvgPool_0a_7x7')
end_points['AvgPool_0a_7x7'] = net
if not num_classes:
return net, end_points
net = slim.dropout(net, dropout_keep_prob, scope='Dropout_0b')
logits = slim.conv2d(net, num_classes, [1, 1], activation_fn=None,normalizer_fn=None, scope='Conv2d_0c_1x1')
if spatial_squeeze:
logits = tf.squeeze(logits, [1, 2], name='SpatialSqueeze')
end_points['Logits'] = logits
end_points['Predictions'] = prediction_fn(logits, scope='Predictions')
return logits, end_points
inception_v1.default_image_size = 224
inception_v1_arg_scope = inception_utils.inception_arg_scope
2.4.2 InceptionV2
参考论文: Batch Normalization:Accelerating Deep Network Training by Reducing Internal Covariate Shift
BN层:
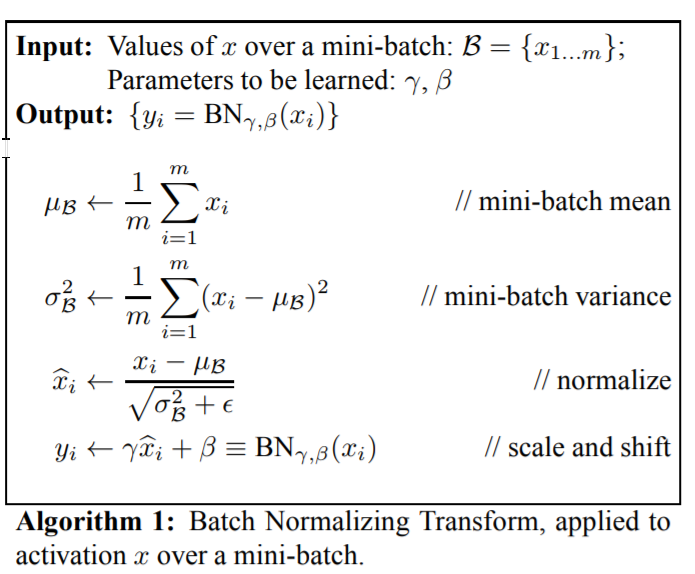
其中,\varepsilon是常数,为数值稳定性添加到小批量方差中。
inception优化的三个模块:

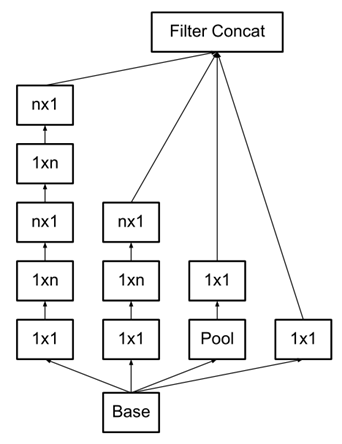
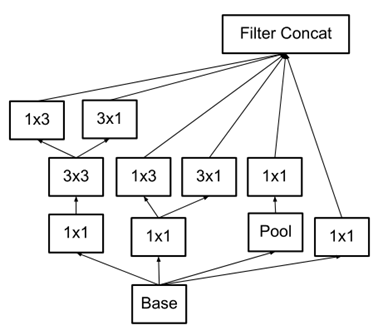
网络结构:

网络特点:
- 使用BatchNorm层代替Dropout层,将每一层的输出都规范化到一个N(0,1)的正态分布,这将有助于训练,因为下一层不必学习输入数据中的偏移,并且可以专注与如何更好地组合特征。
- 使用2个3x3的卷积代替5x5的卷积,这样既可以获得相同的视野(经过2个3x3卷积得到的特征图大小等于1个5x5卷积得到的特征图),还具有更少的参数,提高计算速度,还间接增加了网络的深度
- 将滤波器大小nxn的卷积分解为1xn和nx1卷积的组合。例如,3x3卷积相当于首先执行1x3卷积,然后在其输出上执行3x1卷积。发现这种方法比单个3x3卷积便宜33%。
- 模块中的滤波器组被扩展(更宽而不是更深)以消除代表性瓶颈。如果模块变得更深,则尺寸会过度减少,从而导致信息丢失。
# -*- coding:utf8 -*-
# Contains the definition for inception v2 classification network.
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v1 as tf
import tf_slim as slim
from nets import inception_utils
# pylint: disable=g-long-lambda
trunc_normal = lambda stddev: tf.truncated_normal_initializer(0.0, stddev)
def inception_v2_base(inputs, final_endpoint='Mixed_5c', min_depth=16, depth_multiplier=1.0,
use_separable_conv=True, data_format='NHWC', include_root_block=True, scope=None):
"""Inception v2 (6a2).
Constructs an Inception v2 network from inputs to the given final endpoint.
This method can construct the network up to the layer inception(5b) as
described in http://arxiv.org/abs/1502.03167.
Args:
inputs: a tensor of shape [batch_size, height, width, channels].
final_endpoint: specifies the endpoint to construct the network up to. It
can be one of ['Conv2d_1a_7x7', 'MaxPool_2a_3x3', 'Conv2d_2b_1x1',
'Conv2d_2c_3x3', 'MaxPool_3a_3x3', 'Mixed_3b', 'Mixed_3c', 'Mixed_4a',
'Mixed_4b', 'Mixed_4c', 'Mixed_4d', 'Mixed_4e', 'Mixed_5a', 'Mixed_5b',
'Mixed_5c']. If include_root_block is False, ['Conv2d_1a_7x7',
'MaxPool_2a_3x3', 'Conv2d_2b_1x1', 'Conv2d_2c_3x3', 'MaxPool_3a_3x3'] will
not be available.
min_depth: Minimum depth value (number of channels) for all convolution ops.
Enforced when depth_multiplier < 1, and not an active constraint when
depth_multiplier >= 1.
depth_multiplier: Float multiplier for the depth (number of channels)
for all convolution ops. The value must be greater than zero. Typical
usage will be to set this value in (0, 1) to reduce the number of
parameters or computation cost of the model.
use_separable_conv: Use a separable convolution for the first layer
Conv2d_1a_7x7. If this is False, use a normal convolution instead.
data_format: Data format of the activations ('NHWC' or 'NCHW').
include_root_block: If True, include the convolution and max-pooling layers
before the inception modules. If False, excludes those layers.
scope: Optional variable_scope.
Returns:
tensor_out: output tensor corresponding to the final_endpoint.
end_points: a set of activations for external use, for example summaries or losses.
Raises:
ValueError: if final_endpoint is not set to one of the predefined values, or depth_multiplier <= 0
"""
# end_points will collect relevant activations for external use, for example
# summaries or losses.
end_points = {}
# Used to find thinned depths for each layer.
if depth_multiplier <= 0:
raise ValueError('depth_multiplier is not greater than zero.')
depth = lambda d: max(int(d * depth_multiplier), min_depth)
if data_format != 'NHWC' and data_format != 'NCHW':
raise ValueError('data_format must be either NHWC or NCHW.')
if data_format == 'NCHW' and use_separable_conv:
raise ValueError(
'separable convolution only supports NHWC layout. NCHW data format can'
' only be used when use_separable_conv is False.'
)
concat_dim = 3 if data_format == 'NHWC' else 1
with tf.variable_scope(scope, 'InceptionV2', [inputs]):
with slim.arg_scope( [slim.conv2d, slim.max_pool2d, slim.avg_pool2d],
stride=1, padding='SAME', data_format=data_format):
net = inputs
if include_root_block:
# Note that sizes in the comments below assume an input spatial size of
# 224x224, however, the inputs can be of any size greater 32x32.
# 224 x 224 x 3
end_point = 'Conv2d_1a_7x7'
if use_separable_conv:
# depthwise_multiplier here is different from depth_multiplier.
# depthwise_multiplier determines the output channels of the initial
# depthwise conv (see docs for tf.nn.separable_conv2d), while
# depth_multiplier controls the # channels of the subsequent 1x1
# convolution. Must have
# in_channels * depthwise_multipler <= out_channels
# so that the separable convolution is not overparameterized.
depthwise_multiplier = min(int(depth(64) / 3), 8)
net = slim.separable_conv2d( inputs, depth(64), [7, 7], depth_multiplier=depthwise_multiplier,
stride=2, padding='SAME', weights_initializer=trunc_normal(1.0), scope=end_point)
else:
# Use a normal convolution instead of a separable convolution.
net = slim.conv2d(inputs, depth(64), [7, 7], stride=2, weights_initializer=trunc_normal(1.0), scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint:
return net, end_points
# 112 x 112 x 64
end_point = 'MaxPool_2a_3x3'
net = slim.max_pool2d(net, [3, 3], scope=end_point, stride=2)
end_points[end_point] = net
if end_point == final_endpoint:
return net, end_points
# 56 x 56 x 64
end_point = 'Conv2d_2b_1x1'
net = slim.conv2d(net, depth(64), [1, 1], scope=end_point, weights_initializer=trunc_normal(0.1))
end_points[end_point] = net
if end_point == final_endpoint:
return net, end_points
# 56 x 56 x 64
end_point = 'Conv2d_2c_3x3'
net = slim.conv2d(net, depth(192), [3, 3], scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint:
return net, end_points
# 56 x 56 x 192
end_point = 'MaxPool_3a_3x3'
net = slim.max_pool2d(net, [3, 3], scope=end_point, stride=2)
end_points[end_point] = net
if end_point == final_endpoint:
return net, end_points
# 28 x 28 x 192
# Inception module.
end_point = 'Mixed_3b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(64), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(64), [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(64), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(32), [1, 1], weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 28 x 28 x 256
end_point = 'Mixed_3c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(
net, depth(64), [1, 1],
weights_initializer=trunc_normal(0.09),
scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(96), [3, 3],
scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(
net, depth(64), [1, 1],
weights_initializer=trunc_normal(0.09),
scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3],
scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3],
scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(64), [1, 1], weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 28 x 28 x 320
end_point = 'Mixed_4a'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(128), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_0 = slim.conv2d(branch_0, depth(160), [3, 3], stride=2,scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(64), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(96), [3, 3], scope='Conv2d_0b_3x3')
branch_1 = slim.conv2d(branch_1, depth(96), [3, 3], stride=2, scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.max_pool2d(net, [3, 3], stride=2, scope='MaxPool_1a_3x3')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 14 x 14 x 576
end_point = 'Mixed_4b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(224), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(64), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(96), [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(96), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(128), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(128), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(128), [1, 1], weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 14 x 14 x 576
end_point = 'Mixed_4c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(96), [1, 1],weights_initializer=trunc_normal(0.09),scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(128), [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(96), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(128), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(128), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(128), [1, 1], weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 14 x 14 x 576
end_point = 'Mixed_4d'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(160), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(128), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(160), [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(128), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(160), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(160), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(96), [1, 1], weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 14 x 14 x 576
end_point = 'Mixed_4e'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(96), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(128), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(192), [3, 3],
scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(160), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(192), [3, 3],scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(192), [3, 3],scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(96), [1, 1],weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 14 x 14 x 576
end_point = 'Mixed_5a'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(128), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_0 = slim.conv2d(branch_0, depth(192), [3, 3], stride=2, scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(192), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(256), [3, 3], scope='Conv2d_0b_3x3')
branch_1 = slim.conv2d(branch_1, depth(256), [3, 3], stride=2, scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.max_pool2d(net, [3, 3], stride=2, scope='MaxPool_1a_3x3')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 7 x 7 x 1024
end_point = 'Mixed_5b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(352), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(192), [1, 1],weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(320), [3, 3],scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(160), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(224), [3, 3],scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(224), [3, 3],scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(128), [1, 1],weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 7 x 7 x 1024
end_point = 'Mixed_5c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(352), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(192), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(320), [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(192), [1, 1], weights_initializer=trunc_normal(0.09), scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(224), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(224), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.max_pool2d(net, [3, 3], scope='MaxPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(128), [1, 1], weights_initializer=trunc_normal(0.1), scope='Conv2d_0b_1x1')
net = tf.concat(axis=concat_dim, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
raise ValueError('Unknown final endpoint %s' % final_endpoint)
def inception_v2(inputs, num_classes=1000, is_training=True, dropout_keep_prob=0.8, min_depth=16,depth_multiplier=1.0,
prediction_fn=slim.softmax, spatial_squeeze=True, reuse=None, scope='InceptionV2', global_pool=False):
"""Inception v2 model for classification.
Constructs an Inception v2 network for classification as described in
http://arxiv.org/abs/1502.03167.
The default image size used to train this network is 224x224.
Args:
inputs: a tensor of shape [batch_size, height, width, channels].
num_classes: number of predicted classes. If 0 or None, the logits layer
is omitted and the input features to the logits layer (before dropout)
are returned instead.
is_training: whether is training or not.
dropout_keep_prob: the percentage of activation values that are retained.
min_depth: Minimum depth value (number of channels) for all convolution ops.
Enforced when depth_multiplier < 1, and not an active constraint when
depth_multiplier >= 1.
depth_multiplier: Float multiplier for the depth (number of channels)
for all convolution ops. The value must be greater than zero. Typical
usage will be to set this value in (0, 1) to reduce the number of
parameters or computation cost of the model.
prediction_fn: a function to get predictions out of logits.
spatial_squeeze: if True, logits is of shape [B, C], if false logits is of
shape [B, 1, 1, C], where B is batch_size and C is number of classes.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
scope: Optional variable_scope.
global_pool: Optional boolean flag to control the avgpooling before the
logits layer. If false or unset, pooling is done with a fixed window
that reduces default-sized inputs to 1x1, while larger inputs lead to
larger outputs. If true, any input size is pooled down to 1x1.
Returns:
net: a Tensor with the logits (pre-softmax activations) if num_classes
is a non-zero integer, or the non-dropped-out input to the logits layer
if num_classes is 0 or None.
end_points: a dictionary from components of the network to the corresponding
activation.
Raises:
ValueError: if final_endpoint is not set to one of the predefined values,
or depth_multiplier <= 0
"""
if depth_multiplier <= 0:
raise ValueError('depth_multiplier is not greater than zero.')
# Final pooling and prediction
with tf.variable_scope(scope, 'InceptionV2', [inputs], reuse=reuse) as scope:
with slim.arg_scope([slim.batch_norm, slim.dropout], is_training=is_training):
net, end_points = inception_v2_base( inputs, scope=scope, min_depth=min_depth, depth_multiplier=depth_multiplier)
with tf.variable_scope('Logits'):
if global_pool:
# Global average pooling.
net = tf.reduce_mean(input_tensor=net, axis=[1, 2], keepdims=True, name='global_pool')
end_points['global_pool'] = net
else:
# Pooling with a fixed kernel size.
kernel_size = _reduced_kernel_size_for_small_input(net, [7, 7])
net = slim.avg_pool2d(net, kernel_size, padding='VALID', scope='AvgPool_1a_{}x{}'.format(*kernel_size))
end_points['AvgPool_1a'] = net
if not num_classes:
return net, end_points
# 1 x 1 x 1024
net = slim.dropout(net, keep_prob=dropout_keep_prob, scope='Dropout_1b')
end_points['PreLogits'] = net
logits = slim.conv2d(net, num_classes, [1, 1], activation_fn=None, normalizer_fn=None, scope='Conv2d_1c_1x1')
if spatial_squeeze:
logits = tf.squeeze(logits, [1, 2], name='SpatialSqueeze')
end_points['Logits'] = logits
end_points['Predictions'] = prediction_fn(logits, scope='Predictions')
return logits, end_points
inception_v2.default_image_size = 224
def _reduced_kernel_size_for_small_input(input_tensor, kernel_size):
"""Define kernel size which is automatically reduced for small input.
If the shape of the input images is unknown at graph construction time this
function assumes that the input images are is large enough.
Args:
input_tensor: input tensor of size [batch_size, height, width, channels].
kernel_size: desired kernel size of length 2: [kernel_height, kernel_width]
Returns:
a tensor with the kernel size.
TODO(jrru): Make this function work with unknown shapes. Theoretically, this
can be done with the code below. Problems are two-fold: (1) If the shape was
known, it will be lost. (2) inception.slim.ops._two_element_tuple cannot
handle tensors that define the kernel size.
shape = tf.shape(input_tensor)
return = tf.stack([tf.minimum(shape[1], kernel_size[0]),tf.minimum(shape[2], kernel_size[1])])
"""
shape = input_tensor.get_shape().as_list()
if shape[1] is None or shape[2] is None:
kernel_size_out = kernel_size
else:
kernel_size_out = [min(shape[1], kernel_size[0]),min(shape[2], kernel_size[1])]
return kernel_size_out
inception_v2_arg_scope = inception_utils.inception_arg_scope
2.4.3 InceptionV3
参考论文: Rethinking theInception Architecture for Computer Vision
网络特点:
- RMSProp优化器。
- 学习Factorization into small convolutions的思想,将7x7分解成两个一维的卷积(1x7,7x1),3x3也是一样(1x3,3x1),这样的好处,既可以加速计算(多余的计算能力可以用来加深网络),又可以将1个conv拆成2个conv,使得网络深度进一步增加,增加了网络的非线性,还有值得注意的地方是网络输入从224x224变为了299x299,更加精细设计了35x35/17x17/8x8的模块。
- 标签平滑(添加到损失公式中的一种正规化组件,可防止网络对类过于自信。防止过度拟合)。
# -*- coding:utf8 -*-
# Contains the definition for inception v3 classification network.
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v1 as tf
import tf_slim as slim
from nets import inception_utils
# pylint: disable=g-long-lambda
trunc_normal = lambda stddev: tf.truncated_normal_initializer(0.0, stddev)
def inception_v3_base(inputs, final_endpoint='Mixed_7c', min_depth=16, depth_multiplier=1.0, scope=None):
"""Inception model from http://arxiv.org/abs/1512.00567.
Constructs an Inception v3 network from inputs to the given final endpoint.
This method can construct the network up to the final inception block
Mixed_7c.
Note that the names of the layers in the paper do not correspond to the names
of the endpoints registered by this function although they build the same
network.
Here is a mapping from the old_names to the new names:
Old name | New name
=======================================
conv0 | Conv2d_1a_3x3
conv1 | Conv2d_2a_3x3
conv2 | Conv2d_2b_3x3
pool1 | MaxPool_3a_3x3
conv3 | Conv2d_3b_1x1
conv4 | Conv2d_4a_3x3
pool2 | MaxPool_5a_3x3
mixed_35x35x256a | Mixed_5b
mixed_35x35x288a | Mixed_5c
mixed_35x35x288b | Mixed_5d
mixed_17x17x768a | Mixed_6a
mixed_17x17x768b | Mixed_6b
mixed_17x17x768c | Mixed_6c
mixed_17x17x768d | Mixed_6d
mixed_17x17x768e | Mixed_6e
mixed_8x8x1280a | Mixed_7a
mixed_8x8x2048a | Mixed_7b
mixed_8x8x2048b | Mixed_7c
Args:
inputs: a tensor of size [batch_size, height, width, channels].
final_endpoint: specifies the endpoint to construct the network up to. It
can be one of ['Conv2d_1a_3x3', 'Conv2d_2a_3x3', 'Conv2d_2b_3x3',
'MaxPool_3a_3x3', 'Conv2d_3b_1x1', 'Conv2d_4a_3x3', 'MaxPool_5a_3x3',
'Mixed_5b', 'Mixed_5c', 'Mixed_5d', 'Mixed_6a', 'Mixed_6b', 'Mixed_6c',
'Mixed_6d', 'Mixed_6e', 'Mixed_7a', 'Mixed_7b', 'Mixed_7c'].
min_depth: Minimum depth value (number of channels) for all convolution ops.
Enforced when depth_multiplier < 1, and not an active constraint when
depth_multiplier >= 1.
depth_multiplier: Float multiplier for the depth (number of channels)
for all convolution ops. The value must be greater than zero. Typical
usage will be to set this value in (0, 1) to reduce the number of
parameters or computation cost of the model.
scope: Optional variable_scope.
Returns:
tensor_out: output tensor corresponding to the final_endpoint.
end_points: a set of activations for external use, for example summaries or losses.
Raises:
ValueError: if final_endpoint is not set to one of the predefined values, or depth_multiplier <= 0
"""
# end_points will collect relevant activations for external use, for example
# summaries or losses.
end_points = {}
if depth_multiplier <= 0:
raise ValueError('depth_multiplier is not greater than zero.')
depth = lambda d: max(int(d * depth_multiplier), min_depth)
with tf.variable_scope(scope, 'InceptionV3', [inputs]):
with slim.arg_scope([slim.conv2d, slim.max_pool2d, slim.avg_pool2d], stride=1, padding='VALID'):
# 299 x 299 x 3
end_point = 'Conv2d_1a_3x3'
net = slim.conv2d(inputs, depth(32), [3, 3], stride=2, scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 149 x 149 x 32
end_point = 'Conv2d_2a_3x3'
net = slim.conv2d(net, depth(32), [3, 3], scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 147 x 147 x 32
end_point = 'Conv2d_2b_3x3'
net = slim.conv2d(net, depth(64), [3, 3], padding='SAME', scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 147 x 147 x 64
end_point = 'MaxPool_3a_3x3'
net = slim.max_pool2d(net, [3, 3], stride=2, scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 73 x 73 x 64
end_point = 'Conv2d_3b_1x1'
net = slim.conv2d(net, depth(80), [1, 1], scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 73 x 73 x 80.
end_point = 'Conv2d_4a_3x3'
net = slim.conv2d(net, depth(192), [3, 3], scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 71 x 71 x 192.
end_point = 'MaxPool_5a_3x3'
net = slim.max_pool2d(net, [3, 3], stride=2, scope=end_point)
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# 35 x 35 x 192.
# Inception blocks
with slim.arg_scope([slim.conv2d, slim.max_pool2d, slim.avg_pool2d], stride=1, padding='SAME'):
# mixed: 35 x 35 x 256.
end_point = 'Mixed_5b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(48), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(64), [5, 5], scope='Conv2d_0b_5x5')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(32), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_1: 35 x 35 x 288.
end_point = 'Mixed_5c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(48), [1, 1], scope='Conv2d_0b_1x1')
branch_1 = slim.conv2d(branch_1, depth(64), [5, 5], scope='Conv_1_0c_5x5')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(64), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_2: 35 x 35 x 288.
end_point = 'Mixed_5d'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(48), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(64), [5, 5], scope='Conv2d_0b_5x5')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, depth(96), [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(64), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_3: 17 x 17 x 768.
end_point = 'Mixed_6a'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(384), [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(64), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(96), [3, 3], scope='Conv2d_0b_3x3')
branch_1 = slim.conv2d(branch_1, depth(96), [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_1x1')
with tf.variable_scope('Branch_2'):
branch_2 = slim.max_pool2d(net, [3, 3], stride=2, padding='VALID', scope='MaxPool_1a_3x3')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed4: 17 x 17 x 768.
end_point = 'Mixed_6b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(128), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(128), [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, depth(192), [7, 1], scope='Conv2d_0c_7x1')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(128), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(128), [7, 1], scope='Conv2d_0b_7x1')
branch_2 = slim.conv2d(branch_2, depth(128), [1, 7], scope='Conv2d_0c_1x7')
branch_2 = slim.conv2d(branch_2, depth(128), [7, 1], scope='Conv2d_0d_7x1')
branch_2 = slim.conv2d(branch_2, depth(192), [1, 7], scope='Conv2d_0e_1x7')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(192), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_5: 17 x 17 x 768.
end_point = 'Mixed_6c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(160), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(160), [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, depth(192), [7, 1], scope='Conv2d_0c_7x1')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(160), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(160), [7, 1], scope='Conv2d_0b_7x1')
branch_2 = slim.conv2d(branch_2, depth(160), [1, 7], scope='Conv2d_0c_1x7')
branch_2 = slim.conv2d(branch_2, depth(160), [7, 1], scope='Conv2d_0d_7x1')
branch_2 = slim.conv2d(branch_2, depth(192), [1, 7], scope='Conv2d_0e_1x7')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(192), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_6: 17 x 17 x 768.
end_point = 'Mixed_6d'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(160), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(160), [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, depth(192), [7, 1], scope='Conv2d_0c_7x1')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(160), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(160), [7, 1], scope='Conv2d_0b_7x1')
branch_2 = slim.conv2d(branch_2, depth(160), [1, 7], scope='Conv2d_0c_1x7')
branch_2 = slim.conv2d(branch_2, depth(160), [7, 1], scope='Conv2d_0d_7x1')
branch_2 = slim.conv2d(branch_2, depth(192), [1, 7], scope='Conv2d_0e_1x7')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(192), [1, 1],scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_7: 17 x 17 x 768.
end_point = 'Mixed_6e'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(192), [1, 7],scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, depth(192), [7, 1], scope='Conv2d_0c_7x1')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(192), [7, 1], scope='Conv2d_0b_7x1')
branch_2 = slim.conv2d(branch_2, depth(192), [1, 7], scope='Conv2d_0c_1x7')
branch_2 = slim.conv2d(branch_2, depth(192), [7, 1],
scope='Conv2d_0d_7x1')
branch_2 = slim.conv2d(branch_2, depth(192), [1, 7], scope='Conv2d_0e_1x7')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(192), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_8: 8 x 8 x 1280.
end_point = 'Mixed_7a'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
branch_0 = slim.conv2d(branch_0, depth(320), [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(192), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, depth(192), [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, depth(192), [7, 1], scope='Conv2d_0c_7x1')
branch_1 = slim.conv2d(branch_1, depth(192), [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.max_pool2d(net, [3, 3], stride=2, padding='VALID', scope='MaxPool_1a_3x3')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_9: 8 x 8 x 2048.
end_point = 'Mixed_7b'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(320), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(384), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = tf.concat(axis=3, values=[
slim.conv2d(branch_1, depth(384), [1, 3], scope='Conv2d_0b_1x3'),
slim.conv2d(branch_1, depth(384), [3, 1], scope='Conv2d_0b_3x1')])
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(448), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(384), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = tf.concat(axis=3, values=[
slim.conv2d(branch_2, depth(384), [1, 3], scope='Conv2d_0c_1x3'),
slim.conv2d(branch_2, depth(384), [3, 1], scope='Conv2d_0d_3x1')])
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(192), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
# mixed_10: 8 x 8 x 2048.
end_point = 'Mixed_7c'
with tf.variable_scope(end_point):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, depth(320), [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, depth(384), [1, 1], scope='Conv2d_0a_1x1')
branch_1 = tf.concat(axis=3, values=[
slim.conv2d(branch_1, depth(384), [1, 3], scope='Conv2d_0b_1x3'),
slim.conv2d(branch_1, depth(384), [3, 1], scope='Conv2d_0c_3x1')])
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(net, depth(448), [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, depth(384), [3, 3], scope='Conv2d_0b_3x3')
branch_2 = tf.concat(axis=3, values=[
slim.conv2d(branch_2, depth(384), [1, 3], scope='Conv2d_0c_1x3'),
slim.conv2d(branch_2, depth(384), [3, 1], scope='Conv2d_0d_3x1')])
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(net, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, depth(192), [1, 1], scope='Conv2d_0b_1x1')
net = tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
end_points[end_point] = net
if end_point == final_endpoint: return net, end_points
raise ValueError('Unknown final endpoint %s' % final_endpoint)
def inception_v3(inputs, num_classes=1000, is_training=True, dropout_keep_prob=0.8, min_depth=16, depth_multiplier=1.0,
prediction_fn=slim.softmax, spatial_squeeze=True, reuse=None, create_aux_logits=True, scope='InceptionV3', global_pool=False):
"""Inception model from http://arxiv.org/abs/1512.00567.
"Rethinking the Inception Architecture for Computer Vision"
Christian Szegedy, Vincent Vanhoucke, Sergey Ioffe, Jonathon Shlens,
Zbigniew Wojna.
With the default arguments this method constructs the exact model defined in
the paper. However, one can experiment with variations of the inception_v3
network by changing arguments dropout_keep_prob, min_depth and
depth_multiplier.
The default image size used to train this network is 299x299.
Args:
inputs: a tensor of size [batch_size, height, width, channels].
num_classes: number of predicted classes. If 0 or None, the logits layer
is omitted and the input features to the logits layer (before dropout)
are returned instead.
is_training: whether is training or not.
dropout_keep_prob: the percentage of activation values that are retained.
min_depth: Minimum depth value (number of channels) for all convolution ops.
Enforced when depth_multiplier < 1, and not an active constraint when
depth_multiplier >= 1.
depth_multiplier: Float multiplier for the depth (number of channels)
for all convolution ops. The value must be greater than zero. Typical
usage will be to set this value in (0, 1) to reduce the number of
parameters or computation cost of the model.
prediction_fn: a function to get predictions out of logits.
spatial_squeeze: if True, logits is of shape [B, C], if false logits is of
shape [B, 1, 1, C], where B is batch_size and C is number of classes.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
create_aux_logits: Whether to create the auxiliary logits.
scope: Optional variable_scope.
global_pool: Optional boolean flag to control the avgpooling before the
logits layer. If false or unset, pooling is done with a fixed window
that reduces default-sized inputs to 1x1, while larger inputs lead to
larger outputs. If true, any input size is pooled down to 1x1.
Returns:
net: a Tensor with the logits (pre-softmax activations) if num_classes
is a non-zero integer, or the non-dropped-out input to the logits layer
if num_classes is 0 or None.
end_points: a dictionary from components of the network to the corresponding
activation.
Raises:
ValueError: if 'depth_multiplier' is less than or equal to zero.
"""
if depth_multiplier <= 0:
raise ValueError('depth_multiplier is not greater than zero.')
depth = lambda d: max(int(d * depth_multiplier), min_depth)
with tf.variable_scope(scope, 'InceptionV3', [inputs], reuse=reuse) as scope:
with slim.arg_scope([slim.batch_norm, slim.dropout],is_training=is_training):
net, end_points = inception_v3_base(inputs, scope=scope, min_depth=min_depth, depth_multiplier=depth_multiplier)
# Auxiliary Head logits
if create_aux_logits and num_classes:
with slim.arg_scope([slim.conv2d, slim.max_pool2d, slim.avg_pool2d], stride=1, padding='SAME'):
aux_logits = end_points['Mixed_6e']
with tf.variable_scope('AuxLogits'):
aux_logits = slim.avg_pool2d(aux_logits, [5, 5], stride=3, padding='VALID',scope='AvgPool_1a_5x5')
aux_logits = slim.conv2d(aux_logits, depth(128), [1, 1], scope='Conv2d_1b_1x1')
# Shape of feature map before the final layer.
kernel_size = _reduced_kernel_size_for_small_input(aux_logits, [5, 5])
aux_logits = slim.conv2d(aux_logits, depth(768), kernel_size, weights_initializer=trunc_normal(0.01),
padding='VALID', scope='Conv2d_2a_{}x{}'.format(*kernel_size))
aux_logits = slim.conv2d(aux_logits, num_classes, [1, 1], activation_fn=None,
normalizer_fn=None, weights_initializer=trunc_normal(0.001),scope='Conv2d_2b_1x1')
if spatial_squeeze:
aux_logits = tf.squeeze(aux_logits, [1, 2], name='SpatialSqueeze')
end_points['AuxLogits'] = aux_logits
# Final pooling and prediction
with tf.variable_scope('Logits'):
if global_pool:
# Global average pooling.
net = tf.reduce_mean(input_tensor=net, axis=[1, 2], keepdims=True, name='GlobalPool')
end_points['global_pool'] = net
else:
# Pooling with a fixed kernel size.
kernel_size = _reduced_kernel_size_for_small_input(net, [8, 8])
net = slim.avg_pool2d(net, kernel_size, padding='VALID', scope='AvgPool_1a_{}x{}'.format(*kernel_size))
end_points['AvgPool_1a'] = net
if not num_classes:
return net, end_points
# 1 x 1 x 2048
net = slim.dropout(net, keep_prob=dropout_keep_prob, scope='Dropout_1b')
end_points['PreLogits'] = net
# 2048
logits = slim.conv2d(net, num_classes, [1, 1], activation_fn=None, normalizer_fn=None, scope='Conv2d_1c_1x1')
if spatial_squeeze:
logits = tf.squeeze(logits, [1, 2], name='SpatialSqueeze')
# 1000
end_points['Logits'] = logits
end_points['Predictions'] = prediction_fn(logits, scope='Predictions')
return logits, end_points
inception_v3.default_image_size = 299
def _reduced_kernel_size_for_small_input(input_tensor, kernel_size):
"""Define kernel size which is automatically reduced for small input.
If the shape of the input images is unknown at graph construction time this
function assumes that the input images are is large enough.
Args:
input_tensor: input tensor of size [batch_size, height, width, channels].
kernel_size: desired kernel size of length 2: [kernel_height, kernel_width]
Returns:
a tensor with the kernel size.
TODO(jrru): Make this function work with unknown shapes. Theoretically, this
can be done with the code below. Problems are two-fold: (1) If the shape was
known, it will be lost. (2) inception.slim.ops._two_element_tuple cannot
handle tensors that define the kernel size.
shape = tf.shape(input_tensor)
return = tf.stack([tf.minimum(shape[1], kernel_size[0]),tf.minimum(shape[2], kernel_size[1])])
"""
shape = input_tensor.get_shape().as_list()
if shape[1] is None or shape[2] is None:
kernel_size_out = kernel_size
else:
kernel_size_out = [min(shape[1], kernel_size[0]), min(shape[2], kernel_size[1])]
return kernel_size_out
inception_v3_arg_scope = inception_utils.inception_arg_scope
2.4.4 InceptionV4
参考论文: Inception-ResNet and the Impact of Residual Connections on Learning
stem模块:

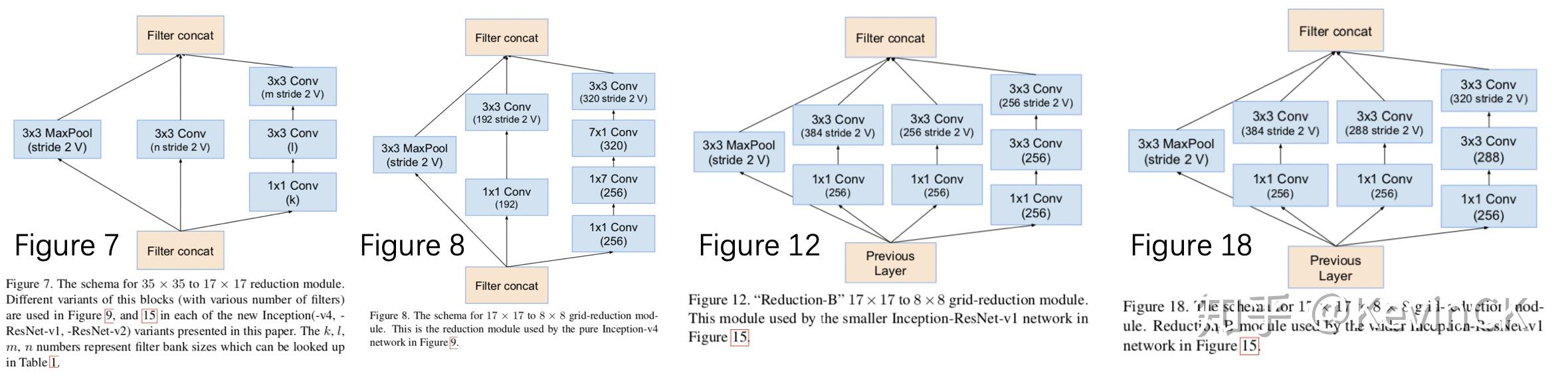
网络图:
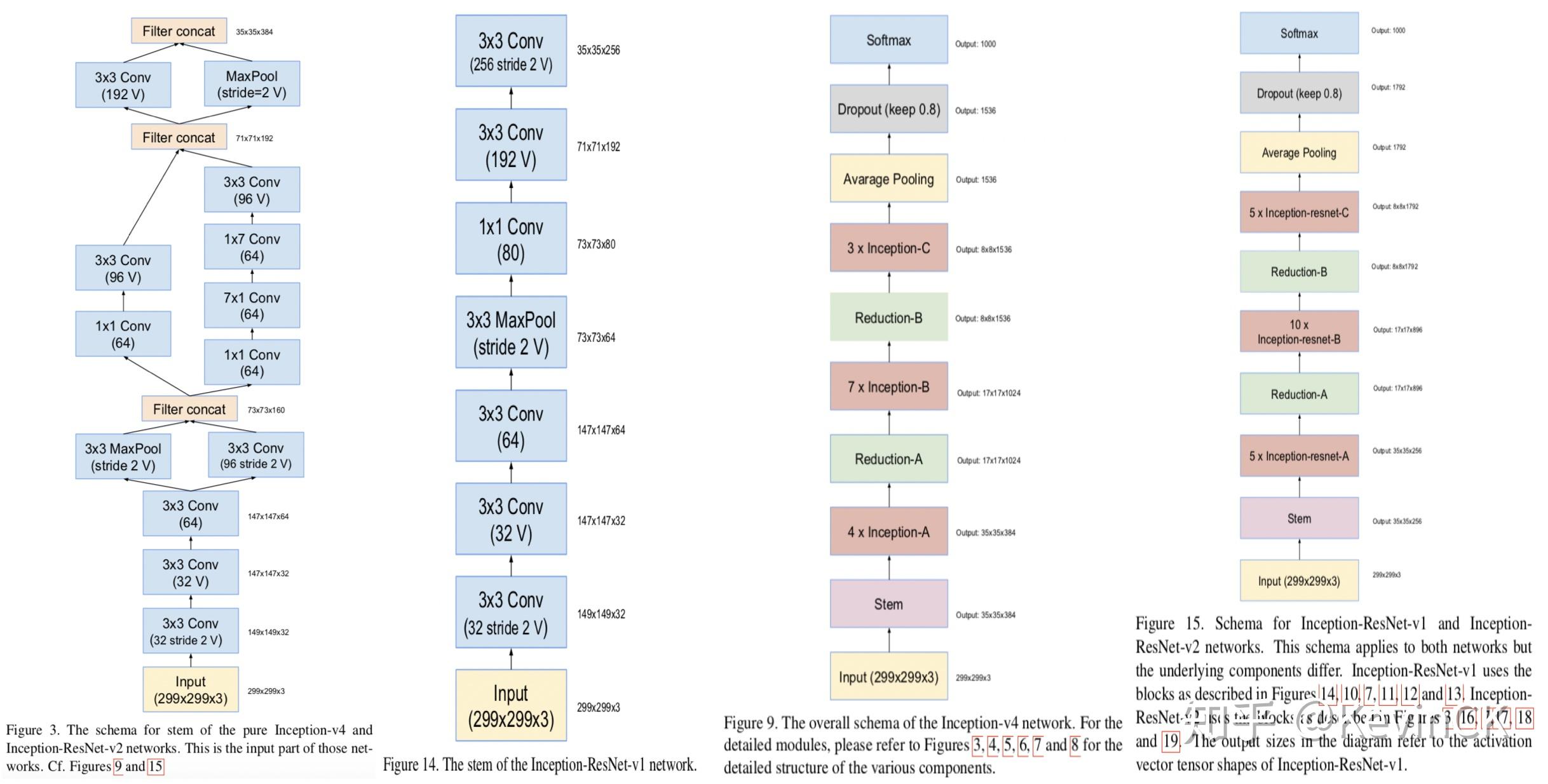
Inception-ResNet V1网络:
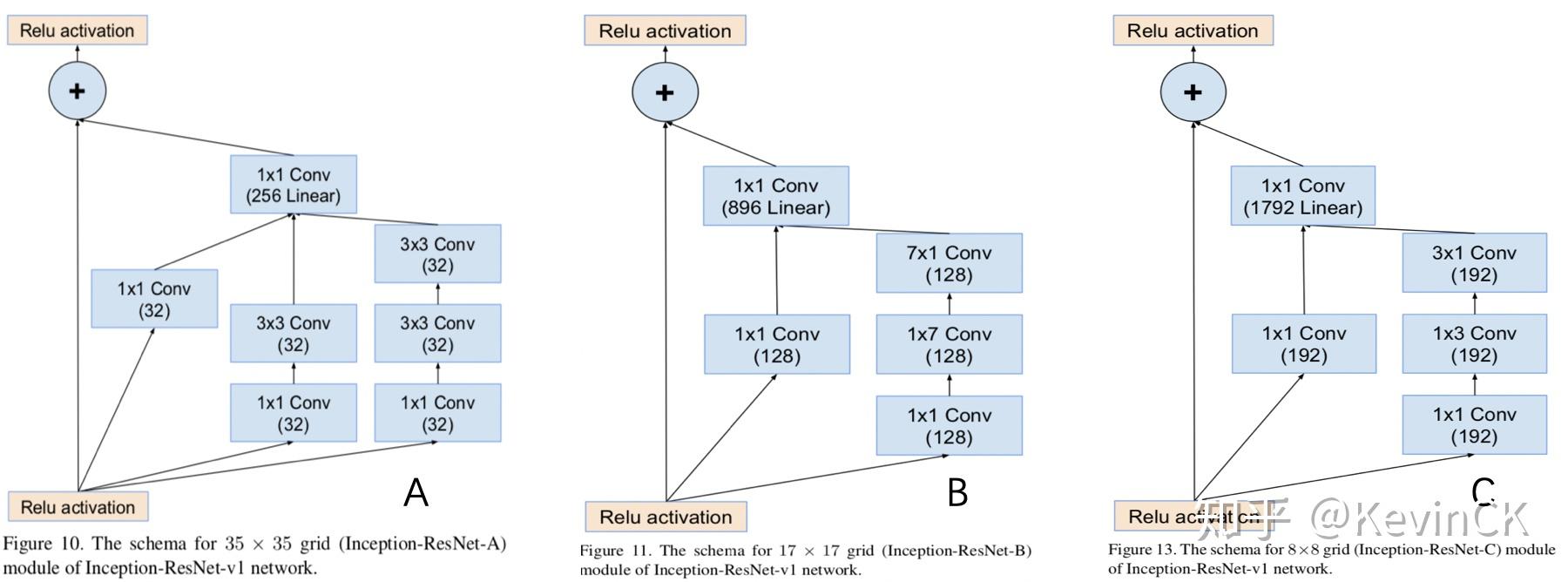
Inception-ResNet V2网络:
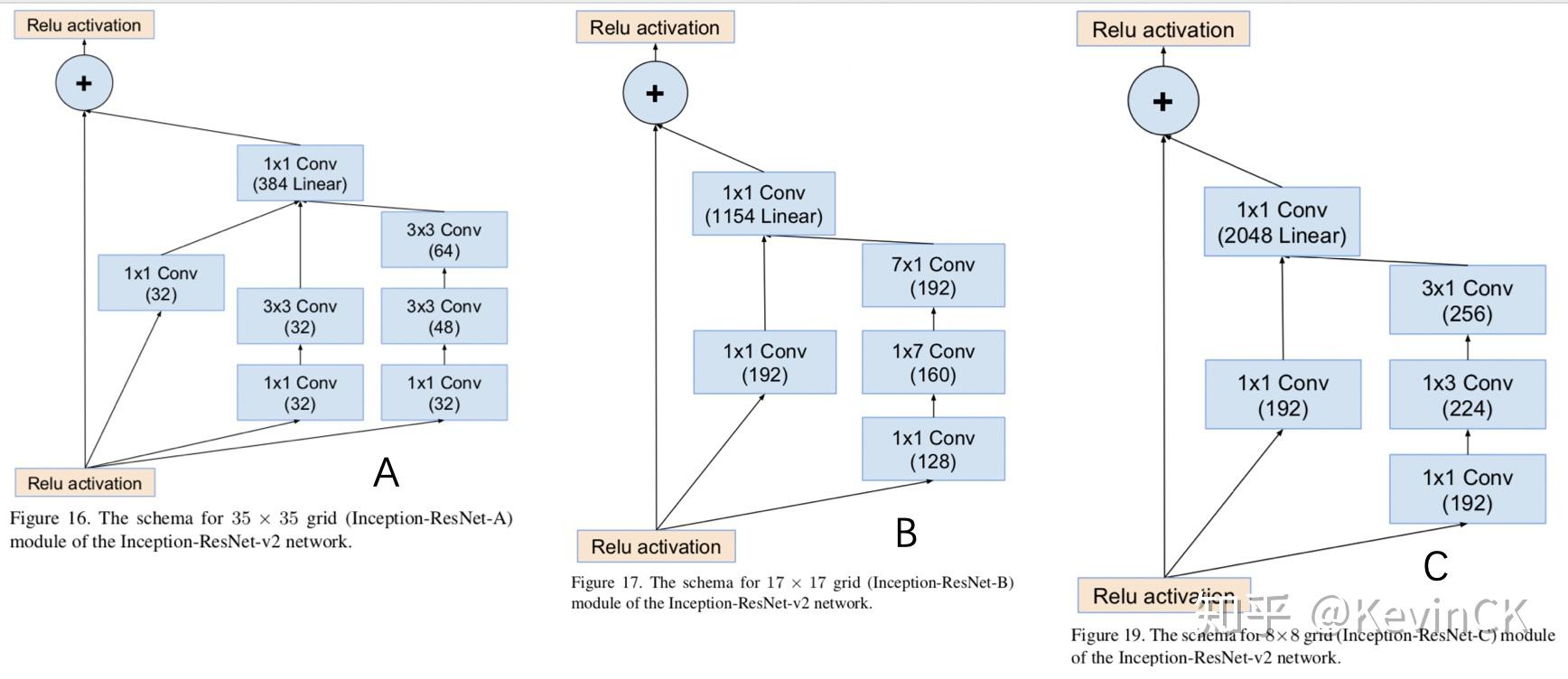
InceptionV4网络特点:
- 修改了stem,这里的stem是指在引入Inception块之前执行的初始操作集。
- V4引入了专门的"Reduction Blocks”,用于改变网格的宽度和高度。早期版本没有显式Reduction Blocks,但实现了类似功能。
- V4版本的3种Inception与之前版本的3种Inception非常相似,但也做了细节的修改。
Inception-ResNet V1和V2特点:
- 在Inception设计中加入了ResNet思想。
- Inception-ResNet V1与V2的整体结构不同,Inception-ResNet V1的计算成本与Inception v3类似,Inception-ResNet V2的计算成本与Inception v4类似。
- 为了实现残差加法,卷积后的输入和输出必须具有相同的尺寸。因此,我们在Inception卷积之后使用1x1卷积来匹配深度大小(卷积后的深度增加)。
- 主要Inception模块内的池化操作被替换为有利于残差连接。但是,您仍然可以在reduction blocks中找到这些操作。Reduction Block A与Inception v4中的相同。
- 如果滤波器的数量超过1000,那么网络架构中更深的残差单元会导致网络"死亡"。因此,为了增加稳定性,作者将残差激活量调整,系数为0.1到0.3。
- 为了在单个GPU上训练模型,原始论文在求和之后没有使用BatchNorm(以使整个模型适合单个GPU)。结果发现,Inception-ResNet模型能够再用较少的epoch时获得更高的精度。
# -*- coding:utf8 -*-
"""
Contains the definition of the Inception V4 architecture.
As described in http://arxiv.org/abs/1602.07261.
Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning
Christian Szegedy, Sergey Ioffe, Vincent Vanhoucke, Alex Alemi
"""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow.compat.v1 as tf
import tf_slim as slim
from nets import inception_utils
def block_inception_a(inputs, scope=None, reuse=None):
# Builds Inception-A block for Inception v4 network.
# By default use stride=1 and SAME padding
with slim.arg_scope([slim.conv2d, slim.avg_pool2d, slim.max_pool2d], stride=1, padding='SAME'):
with tf.variable_scope(scope, 'BlockInceptionA', [inputs], reuse=reuse):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(inputs, 96, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(inputs, 64, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 96, [3, 3], scope='Conv2d_0b_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(inputs, 64, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 96, [3, 3], scope='Conv2d_0b_3x3')
branch_2 = slim.conv2d(branch_2, 96, [3, 3], scope='Conv2d_0c_3x3')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(inputs, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 96, [1, 1], scope='Conv2d_0b_1x1')
return tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
def block_reduction_a(inputs, scope=None, reuse=None):
# Builds Reduction-A block for Inception v4 network.
# By default use stride=1 and SAME padding
with slim.arg_scope([slim.conv2d, slim.avg_pool2d, slim.max_pool2d],stride=1, padding='SAME'):
with tf.variable_scope(scope, 'BlockReductionA', [inputs], reuse=reuse):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(inputs, 384, [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(inputs, 192, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 224, [3, 3], scope='Conv2d_0b_3x3')
branch_1 = slim.conv2d(branch_1, 256, [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.max_pool2d(inputs, [3, 3], stride=2, padding='VALID', scope='MaxPool_1a_3x3')
return tf.concat(axis=3, values=[branch_0, branch_1, branch_2])
def block_inception_b(inputs, scope=None, reuse=None):
# Builds Inception-B block for Inception v4 network.
# By default use stride=1 and SAME padding
with slim.arg_scope([slim.conv2d, slim.avg_pool2d, slim.max_pool2d], stride=1, padding='SAME'):
with tf.variable_scope( scope, 'BlockInceptionB', [inputs], reuse=reuse):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(inputs, 384, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(inputs, 192, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 224, [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, 256, [7, 1], scope='Conv2d_0c_7x1')
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(inputs, 192, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 192, [7, 1], scope='Conv2d_0b_7x1')
branch_2 = slim.conv2d(branch_2, 224, [1, 7], scope='Conv2d_0c_1x7')
branch_2 = slim.conv2d(branch_2, 224, [7, 1], scope='Conv2d_0d_7x1')
branch_2 = slim.conv2d(branch_2, 256, [1, 7], scope='Conv2d_0e_1x7')
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(inputs, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 128, [1, 1], scope='Conv2d_0b_1x1')
return tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
def block_reduction_b(inputs, scope=None, reuse=None):
# Builds Reduction-B block for Inception v4 network.
# By default use stride=1 and SAME padding
with slim.arg_scope([slim.conv2d, slim.avg_pool2d, slim.max_pool2d],stride=1, padding='SAME'):
with tf.variable_scope(
scope, 'BlockReductionB', [inputs], reuse=reuse):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(inputs, 192, [1, 1], scope='Conv2d_0a_1x1')
branch_0 = slim.conv2d(branch_0, 192, [3, 3], stride=2,padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(inputs, 256, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 256, [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, 320, [7, 1], scope='Conv2d_0c_7x1')
branch_1 = slim.conv2d(branch_1, 320, [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_2'):
branch_2 = slim.max_pool2d(inputs, [3, 3], stride=2, padding='VALID', scope='MaxPool_1a_3x3')
return tf.concat(axis=3, values=[branch_0, branch_1, branch_2])
def block_inception_c(inputs, scope=None, reuse=None):
# Builds Inception-C block for Inception v4 network.
# By default use stride=1 and SAME padding
with slim.arg_scope([slim.conv2d, slim.avg_pool2d, slim.max_pool2d],stride=1, padding='SAME'):
with tf.variable_scope(scope, 'BlockInceptionC', [inputs], reuse=reuse):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(inputs, 256, [1, 1], scope='Conv2d_0a_1x1')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(inputs, 384, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = tf.concat(axis=3, values=[
slim.conv2d(branch_1, 256, [1, 3], scope='Conv2d_0b_1x3'),
slim.conv2d(branch_1, 256, [3, 1], scope='Conv2d_0c_3x1')])
with tf.variable_scope('Branch_2'):
branch_2 = slim.conv2d(inputs, 384, [1, 1], scope='Conv2d_0a_1x1')
branch_2 = slim.conv2d(branch_2, 448, [3, 1], scope='Conv2d_0b_3x1')
branch_2 = slim.conv2d(branch_2, 512, [1, 3], scope='Conv2d_0c_1x3')
branch_2 = tf.concat(axis=3, values=[
slim.conv2d(branch_2, 256, [1, 3], scope='Conv2d_0d_1x3'),
slim.conv2d(branch_2, 256, [3, 1], scope='Conv2d_0e_3x1')])
with tf.variable_scope('Branch_3'):
branch_3 = slim.avg_pool2d(inputs, [3, 3], scope='AvgPool_0a_3x3')
branch_3 = slim.conv2d(branch_3, 256, [1, 1], scope='Conv2d_0b_1x1')
return tf.concat(axis=3, values=[branch_0, branch_1, branch_2, branch_3])
def inception_v4_base(inputs, final_endpoint='Mixed_7d', scope=None):
"""
Creates the Inception V4 network up to the given final endpoint.
Args:
inputs: a 4-D tensor of size [batch_size, height, width, 3].
final_endpoint: specifies the endpoint to construct the network up to.
It can be one of [ 'Conv2d_1a_3x3', 'Conv2d_2a_3x3', 'Conv2d_2b_3x3',
'Mixed_3a', 'Mixed_4a', 'Mixed_5a', 'Mixed_5b', 'Mixed_5c', 'Mixed_5d',
'Mixed_5e', 'Mixed_6a', 'Mixed_6b', 'Mixed_6c', 'Mixed_6d', 'Mixed_6e',
'Mixed_6f', 'Mixed_6g', 'Mixed_6h', 'Mixed_7a', 'Mixed_7b', 'Mixed_7c',
'Mixed_7d']
scope: Optional variable_scope.
Returns:
logits: the logits outputs of the model.
end_points: the set of end_points from the inception model.
Raises:
ValueError: if final_endpoint is not set to one of the predefined values,
"""
end_points = {}
def add_and_check_final(name, net):
end_points[name] = net
return name == final_endpoint
with tf.variable_scope(scope, 'InceptionV4', [inputs]):
with slim.arg_scope([slim.conv2d, slim.max_pool2d, slim.avg_pool2d], stride=1, padding='SAME'):
# 299 x 299 x 3
net = slim.conv2d(inputs, 32, [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
if add_and_check_final('Conv2d_1a_3x3', net): return net, end_points
# 149 x 149 x 32
net = slim.conv2d(net, 32, [3, 3], padding='VALID',scope='Conv2d_2a_3x3')
if add_and_check_final('Conv2d_2a_3x3', net): return net, end_points
# 147 x 147 x 32
net = slim.conv2d(net, 64, [3, 3], scope='Conv2d_2b_3x3')
if add_and_check_final('Conv2d_2b_3x3', net): return net, end_points
# 147 x 147 x 64
with tf.variable_scope('Mixed_3a'):
with tf.variable_scope('Branch_0'):
branch_0 = slim.max_pool2d(net, [3, 3], stride=2, padding='VALID', scope='MaxPool_0a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 96, [3, 3], stride=2, padding='VALID', scope='Conv2d_0a_3x3')
net = tf.concat(axis=3, values=[branch_0, branch_1])
if add_and_check_final('Mixed_3a', net): return net, end_points
# 73 x 73 x 160
with tf.variable_scope('Mixed_4a'):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 64, [1, 1], scope='Conv2d_0a_1x1')
branch_0 = slim.conv2d(branch_0, 96, [3, 3], padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.conv2d(net, 64, [1, 1], scope='Conv2d_0a_1x1')
branch_1 = slim.conv2d(branch_1, 64, [1, 7], scope='Conv2d_0b_1x7')
branch_1 = slim.conv2d(branch_1, 64, [7, 1], scope='Conv2d_0c_7x1')
branch_1 = slim.conv2d(branch_1, 96, [3, 3], padding='VALID', scope='Conv2d_1a_3x3')
net = tf.concat(axis=3, values=[branch_0, branch_1])
if add_and_check_final('Mixed_4a', net): return net, end_points
# 71 x 71 x 192
with tf.variable_scope('Mixed_5a'):
with tf.variable_scope('Branch_0'):
branch_0 = slim.conv2d(net, 192, [3, 3], stride=2, padding='VALID', scope='Conv2d_1a_3x3')
with tf.variable_scope('Branch_1'):
branch_1 = slim.max_pool2d(net, [3, 3], stride=2, padding='VALID', scope='MaxPool_1a_3x3')
net = tf.concat(axis=3, values=[branch_0, branch_1])
if add_and_check_final('Mixed_5a', net): return net, end_points
# 35 x 35 x 384
# 4 x Inception-A blocks
for idx in range(4):
block_scope = 'Mixed_5' + chr(ord('b') + idx)
net = block_inception_a(net, block_scope)
if add_and_check_final(block_scope, net): return net, end_points
# 35 x 35 x 384
# Reduction-A block
net = block_reduction_a(net, 'Mixed_6a')
if add_and_check_final('Mixed_6a', net): return net, end_points
# 17 x 17 x 1024
# 7 x Inception-B blocks
for idx in range(7):
block_scope = 'Mixed_6' + chr(ord('b') + idx)
net = block_inception_b(net, block_scope)
if add_and_check_final(block_scope, net): return net, end_points
# 17 x 17 x 1024
# Reduction-B block
net = block_reduction_b(net, 'Mixed_7a')
if add_and_check_final('Mixed_7a', net): return net, end_points
# 8 x 8 x 1536
# 3 x Inception-C blocks
for idx in range(3):
block_scope = 'Mixed_7' + chr(ord('b') + idx)
net = block_inception_c(net, block_scope)
if add_and_check_final(block_scope, net): return net, end_points
raise ValueError('Unknown final endpoint %s' % final_endpoint)
def inception_v4(inputs, num_classes=1001, is_training=True,dropout_keep_prob=0.8, reuse=None, scope='InceptionV4', create_aux_logits=True):
"""Creates the Inception V4 model.
Args:
inputs: a 4-D tensor of size [batch_size, height, width, 3].
num_classes: number of predicted classes. If 0 or None, the logits layer
is omitted and the input features to the logits layer (before dropout)
are returned instead.
is_training: whether is training or not.
dropout_keep_prob: float, the fraction to keep before final layer.
reuse: whether or not the network and its variables should be reused. To be
able to reuse 'scope' must be given.
scope: Optional variable_scope.
create_aux_logits: Whether to include the auxiliary logits.
Returns:
net: a Tensor with the logits (pre-softmax activations) if num_classes
is a non-zero integer, or the non-dropped input to the logits layer
if num_classes is 0 or None.
end_points: the set of end_points from the inception model.
"""
end_points = {}
with tf.variable_scope(scope, 'InceptionV4', [inputs], reuse=reuse) as scope:
with slim.arg_scope([slim.batch_norm, slim.dropout],is_training=is_training):
net, end_points = inception_v4_base(inputs, scope=scope)
with slim.arg_scope([slim.conv2d, slim.max_pool2d, slim.avg_pool2d], stride=1, padding='SAME'):
# Auxiliary Head logits
if create_aux_logits and num_classes:
with tf.variable_scope('AuxLogits'):
# 17 x 17 x 1024
aux_logits = end_points['Mixed_6h']
aux_logits = slim.avg_pool2d(aux_logits, [5, 5], stride=3,padding='VALID',scope='AvgPool_1a_5x5')
aux_logits = slim.conv2d(aux_logits, 128, [1, 1],scope='Conv2d_1b_1x1')
aux_logits = slim.conv2d(aux_logits, 768, aux_logits.get_shape()[1:3], padding='VALID', scope='Conv2d_2a')
aux_logits = slim.flatten(aux_logits)
aux_logits = slim.fully_connected(aux_logits, num_classes,activation_fn=None,scope='Aux_logits')
end_points['AuxLogits'] = aux_logits
# Final pooling and prediction
# TODO(sguada,arnoegw): Consider adding a parameter global_pool which
# can be set to False to disable pooling here (as in resnet_*()).
with tf.variable_scope('Logits'):
# 8 x 8 x 1536
kernel_size = net.get_shape()[1:3]
if kernel_size.is_fully_defined():
net = slim.avg_pool2d(net, kernel_size, padding='VALID', scope='AvgPool_1a')
else:
net = tf.reduce_mean(input_tensor=net, axis=[1, 2],keepdims=True, name='global_pool')
end_points['global_pool'] = net
if not num_classes:
return net, end_points
# 1 x 1 x 1536
net = slim.dropout(net, dropout_keep_prob, scope='Dropout_1b')
net = slim.flatten(net, scope='PreLogitsFlatten')
end_points['PreLogitsFlatten'] = net
# 1536
logits = slim.fully_connected(net, num_classes, activation_fn=None, scope='Logits')
end_points['Logits'] = logits
end_points['Predictions'] = tf.nn.softmax(logits, name='Predictions')
return logits, end_points
inception_v4.default_image_size = 299
inception_v4_arg_scope = inception_utils.inception_arg_scope
https://zhuanlan.zhihu.com/p/66215918
四、检测网络
4.1 Selective Search
- Felzenszwalb算法
参考文章:Efficient Graph-Based Image Segmentation
参考代码: http://cs.brown.edu/people/pfelzens/segment/
Felzenszwalb算法是经典的图像分割算法,是基于图的贪心聚类算法,实现简单,速度比较快,精度也还行。
因为该算法是将图像用加权图抽象化表示,所以补充图的一些基本概念。
图:是由顶点集V(vertices)和边集E(edges)组成,表示为G=(V, E),顶点v∈V,在论文即为单个的像素点,连接一对顶点的边(vi,vj)具有权重w(vi,vj),本文中的意义为顶点之间的不相似度(dissimilarity),所用的是无向图。
树 :特殊的图,图中任意两个顶点,都有路径相连接,但是没有回路。如下图中加粗的边所连接而成的图。如果看成一团乱连的珠子,只保留树中的珠子和连线,那么随便选个珠子,都能把这棵树中所有的珠子都提起来。如果顶点i和h这条边也保留下来,那么顶点h,i,c,f,g就构成了一个回路。
最小生成树(minimum spanning tree) :特殊的树,给定需要连接的顶点,选择边权之和最小的树。
论文中,初始化时每一个像素点都是一个顶点,然后逐渐合并得到一个区域,确切地说是连接这个区域中的像素点的一个MST。如下图,棕色圆圈为顶点,线段为边,合并棕色顶点所生成的MST,对应的就是一个分割区域。分割后的结果其实就是森林。
既然是聚类算法,那应该依据何种规则判定何时该合二为一,何时该继续划清界限呢?对于孤立的两个像素点,所不同的是灰度值,自然就用灰度的距离来衡量两点的相似性,文章中是使用RGB的距离,即
当然也可以用perceptually uniform的Luv或者Lab色彩空间,对于灰度图像就只能使用亮度值了,此外,还可以先使用纹理特征滤波,再计算距离,比如先做Census Transform再计算Hamming distance距离。
对于两个区域(子图)或者一个区域和一个像素点的相似性,最简单的方法即只考虑连接二者的边的不相似度。如下图,已经形成了棕色和绿色两个区域,现在通过紫色边来判断这两个区域是否合并。那么我们就可以设定一个阈值,当两个像素之间的差异(即不相似度)小于该值时,合二为一。迭代合并,最终就会合并成一个个区域,效果类似于区域生长。
对于上右图,显然应该聚成上左图所示的3类:高频区h,斜坡区s,平坦区p。如果我们设置一个全局阈值,那么如果h区要合并成一块的话,那么该阈值要选很大,但是那样就会把p和s区域也包含进来,分割结果太粗。如果以p为参考,那么阈值应该选特别小的值,那样的话p区是会合并成一块,但是h区就会合并成特别特别多的小块,如同一面支离破碎的镜子,分割结果太细。显然,全局阈值并不合适,那么自然就得用自适应阈值。对于p区该阈值要特别小,s区稍大,h区巨大。
先来两个定义,原文依据这两个附加信息来得到自适应阈值。
一个区域内的类内差异Int(C):Int(C)=\max_{e\in(MST,E)}(e),可以近似理解为一个区域内部最大的亮度差异值,定义是MST中不相似度最大的一条边。
两个区域的类间差异Diff(C1, C2):Diff(C_1,C_2)=\min_{v_1\in C_1,v_2\in C_2, (v_1,v_2)\in E}w(v_i,v_j),即连接两个区域所有边中,不相似度最小的边的不相似度,也就是两个区域最相似的地方的不相似度。
直观的判断,当:Diff(C_1,C_2)<min(Int(C_1),Int(C_2))时,两个区域应当合并!
算法步骤:
1 计算每一个像素点与其8邻域或4邻域的不相似度。
如上图,实线为只计算4领域,加上虚线就是计算8邻域,由于是无向图,按照从左到右,从上到下的顺序计算的话,只需要计算右图中灰色的线即可。
2 将边按照不相似度non-decreasing排列(从小到大)排序得到e1,e2, ...,en。
3 选择ei
4 对当前选择的边ej(vi和vj不属于一个区域)进行合并判断。设其所连接的顶点为(vi, vj),
if 不相似度小于二者内部不相似度: 更新阈值以及类标号; else: 如果i < n,则按照排好的顺序,选择下一条边转到Step 4,否则结束。
import cv2
import numpy as np
from skimage import io as sio
from skimage.segmentation import felzenszwalb
import matplotlib.pyplot as plt
from _felzenszwalb_cy import _felzenszwalb_cython # skimage felzenszwalb算法
def felzenszwalb_test(img,sigma,kernel,k, min_size):
# 先使用纹理特征滤波,再计算距离
img = np.asanyarray(img, dtype=np.float) / 255
# rescale scale to behave like in reference implementation
k = float(k) / 255.
img = cv2.GaussianBlur(img, (kernel, kernel), sigma)
height, width = img.shape[:2]
num = height * width
edges = np.zeros(((height - 1) * width * 2 + height * (width - 1) * 2, 3))
# 使用RGB距离,计算四邻域
index = np.array([i for i in range(height * width)])
index = index.reshape((height, width))
to_left = np.sqrt(((img[:, 1:] - img[:, :-1]) ** 2).sum(axis=2))
to_right = to_left
to_up = np.sqrt(((img[1:] - img[:-1]) ** 2).sum(axis=2))
to_down = to_up
last, cur = 0, 0
last, cur = cur, cur + (width - 1) * height
edges[last: cur, 0] = index[:, 1:].reshape(-1)
edges[last: cur, 1] = index[:, :-1].reshape(-1)
edges[last: cur, 2] = to_left.reshape(-1)
last, cur = cur, cur + (width - 1) * height
edges[last: cur, 0] = index[:, :-1].reshape(-1)
edges[last: cur, 1] = index[:, 1:].reshape(-1)
edges[last: cur, 2] = to_right.reshape(-1)
last, cur = cur, cur + (height - 1) * width
edges[last: cur, 0] = index[1:].reshape(-1)
edges[last: cur, 1] = index[:-1].reshape(-1)
edges[last: cur, 2] = to_up.reshape(-1)
last, cur = cur, cur + (height - 1) * width
edges[last: cur, 0] = index[:-1].reshape(-1)
edges[last: cur, 1] = index[1:].reshape(-1)
edges[last: cur, 2] = to_down.reshape(-1)
# 将边按照不相似度从小到大排序
edges = [edges[i] for i in range(edges.shape[0])]
edges.sort(key=lambda x: x[2])
# 构建无向图(树)
class universe(object):
def __init__(self, n, k):
self.f = np.array([i for i in range(n)]) # 树
self.r = np.zeros_like(self.f) # root
self.s = np.ones((n)) # 存储像素点的个数
self.t = np.ones((n)) * k # 存储不相似度
self.k = k
def find(self, x): # Find root of node x
if x == self.f[x]:
return x
return self.find(self.f[x])
def join(self, a, b): # Join two trees containing nodes n and m
if self.r[a] > self.r[b]:
self.f[b] = a
self.s[a] += self.s[b]
else:
self.f[a] = b
self.s[b] += self.s[a]
if self.r[a] == self.r[b]:
self.r[b] += 1
u = universe(num, k)
for edge in edges:
a, b = u.find(int(edge[0])), u.find(int(edge[1]))
if ((a != b) and (edge[2] <= min(u.t[a], u.t[b]))):
# 更新类标号:将的类a,b标号统一为的标号a。更新该类的不相似度阈值为:k / (u.s[a]+u.s[b])
u.join(a, b)
a = u.find(a)
u.t[a] = edge[2] + k / u.s[a]
for edge in edges:
a, b = u.find(int(edge[0])), u.find(int(edge[1]))
if ((a != b) and ((u.s[a] < min_size) or u.s[b] < min_size)):
# 分割后会有很多小区域,当区域像素点的个数小于min_size时,选择与其差异最小的区域合并
u.join(a, b)
dst = np.zeros_like(img)
def locate(index):
return index // width, index % width
avg_color = np.zeros((num, 3))
for i in range(num):
f = u.find(i)
x, y = locate(i)
avg_color[f, :] += img[x, y, :] / u.s[f]
for i in range(height):
for j in range(width):
f = u.find(i * width + j)
dst[i, j, :] = avg_color[f, :]
return dst
if __name__ == '__main__':
sigma = 0.5
kernel = 3
K, min_size = 500, 50
image = sio.imread("test_data/0010.jpg")
# skimage自带的felzenszwalb算法
seg1 = felzenszwalb(image, scale=K, sigma=sigma, min_size=min_size)
# skimage自带的felzenszwalb算法cython版转Python代码,更改了高斯模糊
seg2 = _felzenszwalb_cython(image, scale=K, sigma=sigma, kernel=kernel,min_size=min_size)
# felzenszwalb算法的实现,相比于上一种,区别主要在四邻域和颜色还原
seg3=felzenszwalb_test(image, sigma, kernel,K, min_size)
fig = plt.figure()
a = fig.add_subplot(221)
plt.imshow(image)
a.set_title("image")
a = fig.add_subplot(222)
plt.imshow(seg1)
a.set_title("seg1")
a = fig.add_subplot(223)
plt.imshow(seg2)
a.set_title("seg2")
a = fig.add_subplot(224)
plt.imshow(seg3)
a.set_title("seg3")
plt.show()
- Selective Search
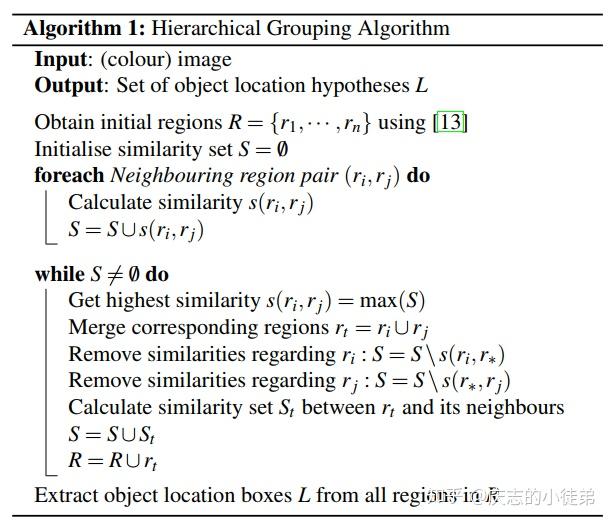
Diversification Strategies:这个部分讲述作者提到的多样性的一些策略,使得抽样多样化,主要有下面三个不同方面:
利用各种不同不变性的色彩空间;
采用不同的相似性度量;
通过改变起始区域。此部分比较简单,不详细介绍,作者对比了一些初始化区域的方法,发现上面效果最好。
Colour Spaces:考虑到场景、光照条件的不同,作者提出使用八种不变性属性的各种颜色空间应用在Hierarchical Grouping Algorithm。如下表:
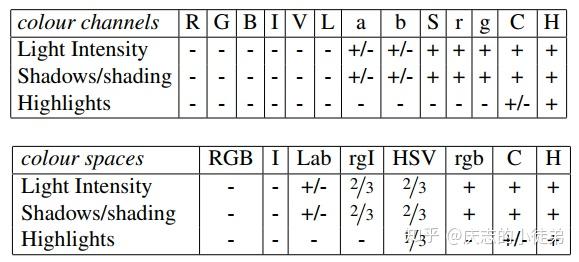
+/-表示部分不变性;1/3表示三个颜色通道中有一个是不变性
Similarity Measures
颜色相似度衡量:C_{i}=\left\{c_{i}^{1}....c_{i}^{n} \right\}表示每一个区域用三通道的颜色直方图表示,每个颜色通道的25 bins的直方图,这样每个区域都可以得到一个n=75维的向量。使用L1-norm标准化后,用下式计算区域间的相似度。
合并区域的颜色直方图计算如下:
纹理相似度衡量: 论文采用SIFT-Like特征,具体操作:采用方差为1的高斯分布对每个颜色通道的8个不同方向做梯度统计,然后将统计结果(尺寸与区域大小一致)以bins=10计算直方图。直方图区间数为8310=240。
T_{i}=\left\{t_{i}^{1}....t_{i}^{n} \right\}表示每一个区域的纹理直方图,有240维。
尺度相似度衡量: 为了保证区域合并操作的尺度较为均匀,使用如下公式,使用尺寸相似度,目的是尽量让小的区域先合并。
size(im)是指区域中的尺寸(以像素为单位)
形状重合度衡量:为了衡量两个区域是否更加重合,合并后区域的Bounding Box越小,其重合度越高,公式如下。
最终的相似度衡量:
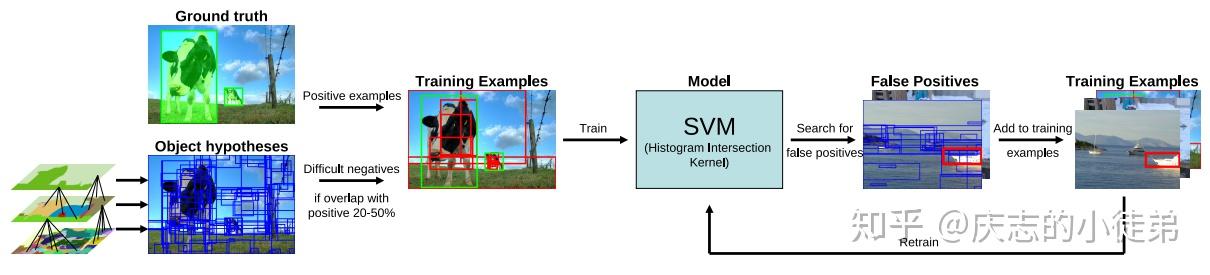
import skimage.io
import skimage.feature
import skimage.color
import skimage.transform
import skimage.util
import skimage.segmentation
import numpy
# "Selective Search for Object Recognition" by J.R.R. Uijlings et al.
#
# - Modified version with LBP extractor for texture vectorization
def _generate_segments(im_orig, scale, sigma, min_size):
"""
segment smallest regions by the algorithm of Felzenswalb and
Huttenlocher
"""
# open the Image
im_mask = skimage.segmentation.felzenszwalb(
skimage.util.img_as_float(im_orig), scale=scale, sigma=sigma,
min_size=min_size)
# merge mask channel to the image as a 4th channel
im_orig = numpy.append(
im_orig, numpy.zeros(im_orig.shape[:2])[:, :, numpy.newaxis], axis=2)
im_orig[:, :, 3] = im_mask
return im_orig
def _sim_colour(r1, r2):
"""
calculate the sum of histogram intersection of colour
"""
return sum([min(a, b) for a, b in zip(r1["hist_c"], r2["hist_c"])])
def _sim_texture(r1, r2):
"""
calculate the sum of histogram intersection of texture
"""
return sum([min(a, b) for a, b in zip(r1["hist_t"], r2["hist_t"])])
def _sim_size(r1, r2, imsize):
"""
calculate the size similarity over the image
"""
return 1.0 - (r1["size"] + r2["size"]) / imsize
def _sim_fill(r1, r2, imsize):
"""
calculate the fill similarity over the image
"""
bbsize = (
(max(r1["max_x"], r2["max_x"]) - min(r1["min_x"], r2["min_x"]))
* (max(r1["max_y"], r2["max_y"]) - min(r1["min_y"], r2["min_y"]))
)
return 1.0 - (bbsize - r1["size"] - r2["size"]) / imsize
def _calc_sim(r1, r2, imsize):
return (_sim_colour(r1, r2) + _sim_texture(r1, r2)
+ _sim_size(r1, r2, imsize) + _sim_fill(r1, r2, imsize))
def _calc_colour_hist(img):
"""
calculate colour histogram for each region
the size of output histogram will be BINS * COLOUR_CHANNELS(3)
number of bins is 25 as same as [uijlings_ijcv2013_draft.pdf]
extract HSV
"""
BINS = 25
hist = numpy.array([])
for colour_channel in (0, 1, 2):
# extracting one colour channel
c = img[:, colour_channel]
# calculate histogram for each colour and join to the result
hist = numpy.concatenate(
[hist] + [numpy.histogram(c, BINS, (0.0, 255.0))[0]])
# L1 normalize
hist = hist / len(img)
return hist
def _calc_texture_gradient(img):
"""
calculate texture gradient for entire image
The original SelectiveSearch algorithm proposed Gaussian derivative
for 8 orientations, but we use LBP instead.
output will be [height(*)][width(*)]
"""
ret = numpy.zeros((img.shape[0], img.shape[1], img.shape[2]))
for colour_channel in (0, 1, 2):
ret[:, :, colour_channel] = skimage.feature.local_binary_pattern(
img[:, :, colour_channel], 8, 1.0)
return ret
def _calc_texture_hist(img):
"""
calculate texture histogram for each region
calculate the histogram of gradient for each colours
the size of output histogram will be
BINS * ORIENTATIONS * COLOUR_CHANNELS(3)
"""
BINS = 10
hist = numpy.array([])
for colour_channel in (0, 1, 2):
# mask by the colour channel
fd = img[:, colour_channel]
# calculate histogram for each orientation and concatenate them all
# and join to the result
hist = numpy.concatenate(
[hist] + [numpy.histogram(fd, BINS, (0.0, 1.0))[0]])
# L1 Normalize
hist = hist / len(img)
return hist
def _extract_regions(img):
R = {}
# get hsv image
hsv = skimage.color.rgb2hsv(img[:, :, :3])
# pass 1: count pixel positions
for y, i in enumerate(img):
for x, (r, g, b, l) in enumerate(i):
# initialize a new region
if l not in R:
R[l] = {
"min_x": 0xffff, "min_y": 0xffff,
"max_x": 0, "max_y": 0, "labels": [l]}
# bounding box
if R[l]["min_x"] > x:
R[l]["min_x"] = x
if R[l]["min_y"] > y:
R[l]["min_y"] = y
if R[l]["max_x"] < x:
R[l]["max_x"] = x
if R[l]["max_y"] < y:
R[l]["max_y"] = y
# pass 2: calculate texture gradient
tex_grad = _calc_texture_gradient(img)
# pass 3: calculate colour histogram of each region
for k, v in list(R.items()):
# colour histogram
masked_pixels = hsv[:, :, :][img[:, :, 3] == k]
R[k]["size"] = len(masked_pixels / 4)
R[k]["hist_c"] = _calc_colour_hist(masked_pixels)
# texture histogram
R[k]["hist_t"] = _calc_texture_hist(tex_grad[:, :][img[:, :, 3] == k])
return R
def _extract_neighbours(regions):
def intersect(a, b):
if (a["min_x"] < b["min_x"] < a["max_x"]
and a["min_y"] < b["min_y"] < a["max_y"]) or (
a["min_x"] < b["max_x"] < a["max_x"]
and a["min_y"] < b["max_y"] < a["max_y"]) or (
a["min_x"] < b["min_x"] < a["max_x"]
and a["min_y"] < b["max_y"] < a["max_y"]) or (
a["min_x"] < b["max_x"] < a["max_x"]
and a["min_y"] < b["min_y"] < a["max_y"]):
return True
return False
R = list(regions.items())
neighbours = []
for cur, a in enumerate(R[:-1]):
for b in R[cur + 1:]:
if intersect(a[1], b[1]):
neighbours.append((a, b))
return neighbours
def _merge_regions(r1, r2):
new_size = r1["size"] + r2["size"]
rt = {
"min_x": min(r1["min_x"], r2["min_x"]),
"min_y": min(r1["min_y"], r2["min_y"]),
"max_x": max(r1["max_x"], r2["max_x"]),
"max_y": max(r1["max_y"], r2["max_y"]),
"size": new_size,
"hist_c": (
r1["hist_c"] * r1["size"] + r2["hist_c"] * r2["size"]) / new_size,
"hist_t": (
r1["hist_t"] * r1["size"] + r2["hist_t"] * r2["size"]) / new_size,
"labels": r1["labels"] + r2["labels"]
}
return rt
def selective_search(
im_orig, scale=1.0, sigma=0.8, min_size=50):
'''Selective Search
Parameters
----------
im_orig : ndarray
Input image
scale : int
Free parameter. Higher means larger clusters in felzenszwalb segmentation.
sigma : float
Width of Gaussian kernel for felzenszwalb segmentation.
min_size : int
Minimum component size for felzenszwalb segmentation.
Returns
-------
img : ndarray
image with region label
region label is stored in the 4th value of each pixel [r,g,b,(region)]
regions : array of dict
[
{
'rect': (left, top, width, height),
'labels': [...],
'size': component_size
},
...
]
'''
assert im_orig.shape[2] == 3, "3ch image is expected"
# load image and get smallest regions
# region label is stored in the 4th value of each pixel [r,g,b,(region)]
img = _generate_segments(im_orig, scale, sigma, min_size)
if img is None:
return None, {}
imsize = img.shape[0] * img.shape[1]
R = _extract_regions(img)
# extract neighbouring information
neighbours = _extract_neighbours(R)
# calculate initial similarities
S = {}
for (ai, ar), (bi, br) in neighbours:
S[(ai, bi)] = _calc_sim(ar, br, imsize)
# hierarchal search
while S != {}:
# get highest similarity
i, j = sorted(S.items(), key=lambda i: i[1])[-1][0]
# merge corresponding regions
t = max(R.keys()) + 1.0
R[t] = _merge_regions(R[i], R[j])
# mark similarities for regions to be removed
key_to_delete = []
for k, v in list(S.items()):
if (i in k) or (j in k):
key_to_delete.append(k)
# remove old similarities of related regions
for k in key_to_delete:
del S[k]
# calculate similarity set with the new region
for k in [a for a in key_to_delete if a != (i, j)]:
n = k[1] if k[0] in (i, j) else k[0]
S[(t, n)] = _calc_sim(R[t], R[n], imsize)
regions = []
for k, r in list(R.items()):
regions.append({
'rect': (
r['min_x'], r['min_y'],
r['max_x'] - r['min_x'], r['max_y'] - r['min_y']),
'size': r['size'],
'labels': r['labels']
})
return img, regions
faster RCNN
https://zhuanlan.zhihu.com/p/156112318
https://zhuanlan.zhihu.com/p/31426458
https://zhuanlan.zhihu.com/p/32404424
https://zhuanlan.zhihu.com/p/112574936
https://zhuanlan.zhihu.com/p/145842317
https://zhuanlan.zhihu.com/p/47579399
https://zhuanlan.zhihu.com/p/145555554
https://zhuanlan.zhihu.com/p/51014564
https://zhuanlan.zhihu.com/p/49082658
https://zhuanlan.zhihu.com/p/62068421
https://zhuanlan.zhihu.com/p/51012194
https://zhuanlan.zhihu.com/p/32404424
https://zhuanlan.zhihu.com/p/60187262
三分支网络
行为识别
https://zhuanlan.zhihu.com/p/45444790
https://zhuanlan.zhihu.com/p/52431409
图像分割
https://zhuanlan.zhihu.com/p/58599382
https://zhuanlan.zhihu.com/p/57674116
CTR相关
https://zhuanlan.zhihu.com/p/57987311
https://zhuanlan.zhihu.com/p/35465875
https://zhuanlan.zhihu.com/p/23499698
https://fuhailin.github.io/GBDT-LR/
https://zhuanlan.zhihu.com/p/60704781
边界网络
https://zhuanlan.zhihu.com/p/75444151
https://zhuanlan.zhihu.com/p/57995509
https://zhuanlan.zhihu.com/p/57988740
https://zhuanlan.zhihu.com/p/65306548